Home
> Musings: Main
> Archive
> Archive for January-April 2013 (this page)
| Introduction
| e-mail announcements
| Contact
Musings: January-April 2013 (archive)
Musings is an informal newsletter mainly highlighting recent science. It is intended as both fun and instructive. Items are posted a few times each week. See the Introduction, listed below, for more information.
If you got here from a search engine... Do a simple text search of this page to find your topic. Searches for a single word (or root) are most likely to work.
Introduction (separate page).
This page:
2013 (January-April)
April 30
April 24
April 17
April 10
April 3
March 27
March 20
March 13
March 6
February 27
February 20
February 13
February 6
January 30
January 23
January 16
January 9
January 3
Also see the complete listing of Musings pages, immediately below.
All pages:
Most recent posts
2026
2025
2024
2023:
January-April
May-December
2022:
January-April
May-August
September-December
2021:
January-April
May-August
September-December
2020:
January-April
May-August
September-December
2019:
January-April
May-August
September-December
2018:
January-April
May-August
September-December
2017:
January-April
May-August
September-December
2016:
January-April
May-August
September-December
2015:
January-April
May-August
September-December
2014:
January-April
May-August
September-December
2013:
January-April: this page, see detail above
May-August
September-December
2012:
January-April
May-August
September-December
2011:
January-April
May-August
September-December
2010:
January-June
July-December
2009
2008
Links to external sites will open in a new window.
Archive items may be edited, to condense them a bit or to update links. Some links may require a subscription for full access, but I try to provide at least one useful open source for most items.
Please let me know of any broken links you find -- on my Musings pages or any of my regular web pages. Personal reports are often the first way I find out about such a problem.
April 30, 2013
Using your brain waves to log on to the computer
April 29, 2013

|
That is UC Berkeley professor John Chuang, wearing a headset that may allow him to log on to his computer simply by thinking of his password -- or "pass-thought". The key part is the arm of the headset that is pressed against a specific region of his forehead: a sensor to capture brain waves from his frontal cortex.
This is from the first news story, below.
|
A news story about this work reminded me of a recent post, in which brain waves were sent from one animal to another [link at the end]. The news story is based on a talk given recently by Chuang's group. Even though the information available is limited, it seemed of interest to briefly note this new work.
Of course, it isn't the idea here that is novel, but the implementation. There are a couple of main themes in this work:
1) One is that they use an inexpensive commercially available device to collect brain waves. It is a single channel electroencephalogram (EEG) device, with a sensor worn on the forehead, as shown in the picture above. The signal -- the brain waves from the person trying to log on -- is compared to what the person has previously recorded; in a sense, it is just like a regular password system, but what is checked is what the person thinks, not what the person types.
This device is much simpler than the complex EEG sensors used when the goal is to assist people with disabilities. In the current work, the goal is relatively simple: they need only associate the brain waves with the individual. When brain waves are being used to perform tasks, a more refined signal is needed.
In fact, one of their conclusions is that they now get results with the simple EEG that are as good as previous results with the more complex "clinical" EEGs.
2) The other theme is what we might call consumer acceptance. Much of the talk is about what kinds of mental tasks people might find appropriate for this computer operation. They find that people vary in what they consider easy or boring. Their general conclusion is that it is probably best to allow users to select their own type of task.
Does it work? With their best system, they get an error rate of about 1% -- mostly due to false rejection of the proper user (rather than to acceptance of an improper user). As noted, an important point for them is that they achieve this with a simple device. They do not claim it is ready for real world use, but rather that it is worth studying further.
News stories:
* New Research: Computers That Can Identify You by Your Thoughts. (UC Berkeley School of Information, April 3, 2013.) Good overview.
* Thoughts could be future of security. (Daily Cal (UC Berkeley student newspaper), April 10, 2013. Now archived.) A more informal overview.
Chuang's group gave a talk about this work recently at a meeting; here is the paper they presented. It is freely available; the link here is to a copy from the authors: I Think, Therefore I Am: Usability and Security of Authentication Using Brainwaves. (J Chuang et al, 2013 Workshop on Usable Security, Seventeenth International Conference on Financial Cryptography and Data Security, April 2013.) Remember, this is a meeting talk, not a published paper. We note it here briefly as fun and interesting -- and potentially useful. And it is, for the most part, a quite readable paper. Formal publication of a peer-reviewed paper will presumably follow at some point.
Background post: Can one rat know what another rat is thinking? (April 8, 2013). A brain signal is sent from one rat to another; the receiving rat acts on the signal.
Examples of the use of brain-computer interface to assist the disabled.
* Brain-computer interface -- without invasive electrodes (December 28, 2016).
* Brain-computer interface: Paralyzed patients control robotic arm by their thoughts (June 16, 2012).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
More about computer security:
* Computer security: web-based password managers (September 29, 2014).
* Can computers talk to each other? Could it be a new type of security threat? (June 18, 2014).
Loudspeakers: From gold-coated pig intestine to graphene
April 27, 2013
Audio devices transmit sound to us via vibrating membranes, driven electrically or magnetically. We refer to the membrane devices with terms such as loudspeakers or headphones. Desired characteristics of such devices include a flat frequency response over the range of human hearing, say from 20 to 20,000 Hertz (Hz), and good power efficiency.
Mankind has been making such devices for decades. Early electrically-driven phones, using gold-coated pig intestine as the material for the vibrating membrane, are long forgotten. Now, UC Berkeley physicists announce that they can do better than high-quality commercial headphones almost on their first try using graphene membranes only 30 nanometers (nm) thick. Why graphene? First, it is naturally electrically conducting. Second, it is incredibly strong, allowing use of a very thin membrane.
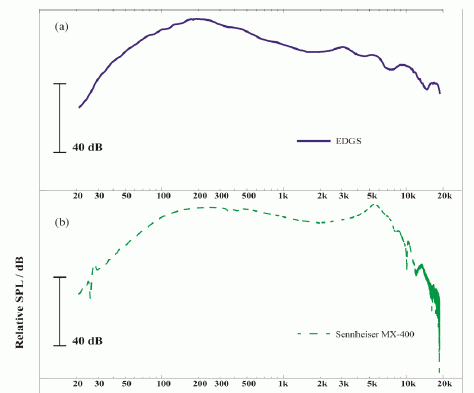
|
Above are frequency response curves for two headphone devices.
The x-axis is the sound frequency, from 20 to 20,000 Hz -- on a log scale. The y-axis shows the headphone response, shown as the relative sound pressure level (SPL), in decibels (dB).
Part a (upper) is for the electrostatically driven graphene speaker (EDGS) -- the device they made. Part b (lower) is for a high-quality commercial headphone.
You can see that the two devices perform similarly for much of the frequency range. At high frequencies, the graphene speaker outperforms the commercial headphone. (The tail-off at low frequencies is probably an artifact of their measurements.) Their subjective observations are in agreement with these data.
This is part of Figure 3 from the article.
|
Their general conclusion is that, with little development effort, they have made headphones of quality similar to if not better than those commonly used. In addition to the good frequency response, their graphene headphones are efficient -- probably 10-fold more efficient than the commercial phones; that's an issue in these days of battery-operated devices.
As noted above, the secret is the strength of graphene, allowing use of a very thin membrane. This simplifies the design. In particular, a thin membrane is damped simply by the surrounding air, without needing a separate damping system. In addition to being simpler, this allows more of the energy to go directly into producing sound. (Damping refers to stopping a vibration. If damping did not occur, one sound would pile upon the previous one.)
Is this going to make it to market? I don't know. There is also a gap between lab-scale and commercial development. But it does sound like it may be worth exploring. Remember, graphene is a rather new material, and people are still learning how to use it.
News stories:
* First Graphene Audio Speaker Easily Outperforms Traditional Designs. (Physics arXiv Blog (MIT Technology Review), March 13, 2013.)
* Experimental graphene earphones outperform most commercial headsets. (Nanowerk, March 21, 2013.) (Caution... The title of this item notes "most" commercial headphones. In fact, they seem to have tested only one. This is a problem of headline writing; the article itself seems quite on-target.)
* UC Berkeley researchers develop first graphene-based headphones. (Daily Cal (UC Berkeley student newspaper), March 18, 2013. Now archived.) Starts with a funny story.
The article: Electrostatic graphene loudspeaker. (Q Zhou & A Zettl, Applied Physics Letters 102:223109, June 5, 2013.) It is also available from Zettl's web site: Zettl publication list -- scroll down to the item.
More on graphene:
* A simple way to make a supercapacitor with high energy storage? (January 6, 2014).
* Graphene bubbles: tiny adjustable lenses? (January 15, 2012).
A post about carbon nanotubes, which are closely related to graphene: Characterization of carbon nanotubes (December 3, 2013).
More about sound: The golden ear: A nano-ear based on optical tweezers (July 13, 2012).
More about "listening": A rapid test for antibiotic sensitivity? (July 19, 2013).
More from Alex Zettl: CITRIS: Zettl; new energy series (November 1, 2009).
This post is listed on my page Introduction to Organic and Biochemistry -- Internet resources in the section on Aromatic compounds.
Hard seeds or soft seeds?
April 26, 2013
Interesting little story. Some plants make hard seeds, some make soft seeds; some make both kinds. Why?
The common view has been that seed hardness provides a physical basis for dormancy, promoting the lifetime of the seeds. A new article suggests another advantage of hard seeds: they may be less attractive to animals -- because they give off less of the odor molecules that attract the predators.
Their experimental approach is simple and fun. The scientists provide hamsters with an array of little dishes filled with various little things... soft seeds, hard seeds, and gravel. (In a given test, the hard and soft seeds are from the same type of plant.) They measure what the hamsters find.

|
A hamster looking for seeds.
This is trimmed and reduced from a figure in the news story. Figure 1 of the paper shows several similar scenes, as well as the general test set-up.
|
In some experiments, the hamsters could see the seeds. In that case, hard vs soft made little difference. However, in some experiments, the seeds were under the gravel, and could be detected only by odor. In that case, the hamsters removed many more soft seeds than hard ones.
The simple conclusion, then, is that hard seeds are harder for the hamsters to detect by odor.
That is, the hard seed is an anti-predator trait.
The authors speculate about the importance of various reasons for the seed types, but there is not much to go on. One particularly interesting speculation is about why a plant might make both hard and soft seeds. The soft seeds attract animals, and the hard ones may go straight through the digestive system. Thus the mixture serves to promote dispersal of the seeds.
A bit more explanation... A key feature of "hard" seeds is that they are impermeable to water. They do not easily germinate -- and start to metabolize -- when exposed to water. Thus the general feature of impermeability, which promotes dormancy, also reduces release of the volatiles that the hamsters detect, probably by preventing their production by metabolism. The key experiments, in which odor detection was important, used seeds that were not only buried but also wetted, to promote metabolism.
News story: Physical dormancy in seeds: a game of hide and seek? (New Phytologist Trust, March 8, 2013.) A useful brief overview.
The article: Physical dormancy in seeds: a game of hide and seek? (T R Paulsen et al, New Phytologist 198:496, April 2013.)
More about seeds: Miniature helicopters -- and botany (July 6, 2009).
More about detecting food by odor:
* Malaria-infected mosquitoes have greater attraction for people (May 28, 2013).
* What happens if you block the left nostril of a mole's nose? (April 19, 2013).
More, May 7, 2013... Comment. A reader questioned a word usage in this post. Is it proper to use the term predator to describe an animal that eats seeds? I had wondered, too, but gone along with the usage by the authors of the paper. Turns out that seed predation is a well-accepted usage; put the term in a search engine or Wikipedia, if you want.
April 24, 2013
It's a dog-eat-starch world
April 23, 2013

|
That's the idea.
The picture is actually a fake -- a composite. Still, it's cute.
This is from the Broad Institute news story.
|
The point, however, is important. Dogs do digest starch -- and that is noteworthy. Dogs are of the carnivore family, descended from wolves. Wolves are rather strict carnivores.
A new article examines the genomes of wolves and dogs. Many of the characteristic differences are in brain genes. That finding is expected, since there are numerous behavioral differences between dogs and wolves. However, another major group of differences is in genes for digestion; several of these differences seem to promote the expansion of the digestive abilities of the dog. For example... One key enzyme for breaking down starch is amylase; dogs contain several times more amylase than wolves do.
There are two broad reasons why this is of interest...
* One is for its insight into the evolution of dogs. It is well accepted that dogs descended from wolves. They adapted to become more sociable; the changes in brain genes presumably relate to this. And they adapted to our diet, the diet of agricultural man. Perhaps genetic adaptation to digesting starch was an early event, as some wolves began to explore the garbage piles left by humans. However, we should stress that the genome data yields little information at this point on the timing of any of these changes; much of what you read about the steps in the evolution of the dog is speculation.
* The other is that the change in dog digestion mirrors a change in human digestion. Humans, too, changed to a more starchy diet with the dawn of agriculture. This change was noted in a recent post [link at end].
News stories:
* Dogs Adapted to Agriculture -- As wolves became domesticated, their genes adapted to a starch-rich diet of human leftovers. (The Scientist, January 23, 2013.)
* In divergence from wolves, doggie diet made a difference. (Broad Institute (MIT & Harvard), January 23, 2013.)
* News story accompanying the article: Evolutionary genomics: Detecting selection. (G S Barsh & L Andersson, Nature 495:325, March 21, 2013.)
* The article: The genomic signature of dog domestication reveals adaptation to a starch-rich diet. (E Axelsson et al, Nature 495:360, March 21, 2013.)
Background post on human digestion: Bacteria on human teeth -- through the ages (March 24, 2013).
More about dogs (and wolves):
* Will a wolf puppy play ball with you? (February 7, 2020).
* The oldest known dog leash? (January 23, 2018).
* Doggy bags and the food waste problem (January 4, 2017).
* Sharing microbes within the family: kids and dogs (May 14, 2013).
* Dog fMRI (June 8, 2012).
* Pet Diary (September 25, 2009).
More about carnivores:
* Loss of ability to taste "sweet" in carnivores (April 6, 2012).
* Carnivorous plants: A blue glow (March 16, 2013).
More about domestication... Atmospheric CO2 and the origin of domesticated corn (February 14, 2014).
There is more about genomes on my page Biotechnology in the News (BITN) - DNA and the genome.
Life in an Antarctic lake
April 22, 2013
Briefly noted...
Lake Vida is a lake in Antarctica. It is covered with ice, and has probably been isolated from external input for nearly 3,000 years. We would consider the conditions in Lake Vida to be rather extreme. The temperature is about -13 °C, and it is quite salty but devoid of oxygen. In a recent article, a team of scientists report that Lake Vida is teeming with life -- microbes.
Exploring the sub-surface lakes of Antarctica is a new and active field. It's also a difficult and controversial field. The idea is to isolate water from these underground lakes -- without contaminating them with anything from the outside world. It involves drilling a hole -- while maintaining absolute biological sterility. And doing it under some of the most inhospitable conditions on Earth.
In recent weeks there have been news stories about possible isolation of novel microbes from an Antarctic lake thought to be isolated for millions of years. The initial news report was followed by denials, and counter-denials. Unfortunately, at this point there are no real facts on that story.
The current article is a small step in the study of Antarctica's underground lakes: a published paper about a younger lake. It opens the subject of finding microbial life in such lakes. Published papers do not always turn out to be right, but at least a formal publication gives us something solid to look at.
The article is largely a description of what they found, both for chemicals and microbes. As examples... The lake water contains 20% salt. It is supersaturated with N2O (nitrous oxide, laughing gas). There is a diverse collection of microbes (about 32 species across eight phyla, based on analyzing DNA), at around 106/mL. It is presumed that the energy source for the microbes is inorganic chemicals, deriving from the rocks in contact with the lake.
News story: Hearty Organisms Discovered in Bitter-Cold Antarctic Brine. (Science Daily, November 26, 2012.) (One might guess that the first word of the title should be "hardy". The "error" apparently derives from a press release from one of the lead institutions involved in the work.) A useful overview.
The article, which is freely available: Microbial life at -13 °C in the brine of an ice-sealed Antarctic lake. (A E Murray et al, PNAS 109:20626, December 11, 2012.)
More from Antarctica:
* What do microbes eat when there is nothing to eat in Antarctica? (April 2, 2018).
* A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
* How were the Gamburtsevs formed? (December 7, 2011).
* How an octopus adapts to the cold -- by RNA editing (March 5, 2012).
Underground water on Mars? ... A lake on Mars? (August 24, 2018).
Melamine toxicity: possible role of gut microbiota
April 21, 2013
A few years ago, there were two well-publicized incidents of poisoning due to the chemical melamine. One involved pet food, and the other involved milk used for infants. These incidents probably involved deliberate adulteration of the food products with melamine. Why? Because melamine is counted as "protein" by common analyses, and it is cheaper than real protein.
But if the incentive was economic, why would anyone adulterate a food with a poison? Because melamine is not poisonous. And thus we begin to see the melamine mystery. Not only were these cases of fraud, but they were also cases of poisoning by something that was not poisonous. There must be more to the story.
Scientists immediately investigated, and soon came to the conclusion that the melamine was toxic by interacting with another chemical, called cyanuric acid (CA); the combination of melamine plus CA led to kidney stones. With CA, melamine was toxic; without CA, melamine was not toxic. It was plausible that the melamine used for adulteration contained some CA (the two chemicals are actually related).
A recent article introduces a new twist to the melamine story. A team of scientists now suggests that animals consuming melamine may convert some of it to CA. More specifically, they suggest -- and show -- that the conversion is done by bacteria in the gut of the animals, that is, by the gut microbiota. If this is correct, then consumption of "pure" melamine might lead to kidney stones of melamine plus CA, because the animals, via their gut bacteria, make the CA.
It was already known that some bacteria can convert melamine to CA. With the increased understanding of the importance of the gut microbiota, scientists wondered if this might be relevant to the melamine poisoning. In their first experiment, they simply tested the effect of giving an antibiotic on melamine toxicity in rats. The antibiotic treatment reduced the melamine toxicity! That alone is an interesting result, and would seem to implicate bacteria -- somehow -- in melamine toxicity.
The scientists then went on to isolate bacteria from the rat feces that could convert melamine to CA. They ended up focusing attention on Klebsiella bacteria, particularly an isolate of Klebsiella terrigena. Here is an example of what they found using this strain...
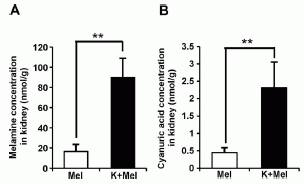
|
In this experiment, rats were given melamine; some were also given the Klebsiella bacteria. The bars are labeled "Mel" for melamine alone, or "K+Mel" for Klebsiella bacteria + melamine.
The bars show the chemicals found in the kidneys -- presumably in the form of kidney stones. The left hand graph (A) is for melamine in the kidneys; the right hand graph (B) is for cyanuric acid (CA).
|
You can see that the levels of both chemicals were enhanced by adding the bacteria. The interpretation is that the bacteria convert some of the melamine to CA, and that promotes kidney stone formation.
This is Figure 5 parts A & B from the article.
|
Overall, we have evidence that bacteria can promote the formation of CA from melamine, and that they can promote kidney stone formation -- presumably by that conversion. This offers one more clue as to how melamine can be toxic.
The story is incomplete, however. If normal gut microbiota make melamine toxic by converting some of it to CA, why then does melamine test as non-toxic? A possible answer is that the prevalence of CA-forming bacteria varies widely. The authors even speculate that the level of such bacteria was one factor determining why some children were more affected by the melamine-containing milk.
Here is another "loose end"... Some work showed that the children who were poisoned by the melamine formed kidney stones with the melamine combined with uric acid, not CA. Interestingly, the experiment described above also showed increased levels of uric acid in the kidneys (part C of the figure, not shown here). The significance is not clear. It is possible that this simply reflects greater stone formation, with uric acid being included in the stones. There is no evidence about whether the bacteria might be stimulating uric acid production. The point for now is that CA may be only part of the story. It is plausible that clinically relevant kidney stones are due to melamine interacting with both CA and uric acid.
News story: Gut Microbes Could Determine the Severity of Melamine-Induced Kidney Disease. (Science Daily, February 14, 2013.)
The article: Melamine-Induced Renal Toxicity Is Mediated by the Gut Microbiota. (X Zheng et al, Science Translational Medicine 5:172ra22, February 13, 2013.)
For more on the melamine story, see my page of Internet resources for Introduction to Organic and Biochemistry, in the section on Amines, amides. That page includes structures of melamine and CA.
For more about the gut microbiota:
* A phage treatment for inflammatory bowel disease (August 13, 2022).
* Malnutrition: is more (or better) food the answer? (March 8, 2013).
* Sharing microbes within the family: kids and dogs (May 14, 2013). A broader perspective.
* Red meat and heart disease: carnitine, your gut bacteria, and TMAO (May 21, 2013).
A post about the bacteria associated with a sponge: Theonella's secret: Entotheonella (March 18, 2014).
More melamine: Artificial wood (November 3, 2018).
For another story about melamine-related chemicals, see A novel type of polymer -- and its possible relevance to the origin of life (March 15, 2013).
More about adulteration: Purity of dietary supplements? (October 23, 2018).
What happens if you block the left nostril of a mole's nose?
April 19, 2013
It veers to the right.

|
Scalopus aquaticus, the common American mole.
Focus on the nose.
This picture is from the Science Daily news story.
|
Humans have two ears, spaced some distance apart. Each ear sends a signal to the brain, which then integrates the two signals to obtain information about the direction and distance of the sound source. This is an example of stereo sensing.
Little is known about smelling in stereo. A new article shows that one animal seems to be able naturally to smell in stereo. It is the mole, shown above. This little burrower depends on smell as its primary sense for finding food.
The work began by noticing that the moles shifted their head back and forth while sniffing out a food source. This suggests that they are processing repeated sniffs to gain information about the source. This observation led a scientist to set up controlled conditions for studying how the moles respond to odor cues. The testing included tests not only for serial sniffing but for stereo sniffing: integration of the distinct information from the two nostrils.
A key piece of evidence came from experiments of the type noted in the title of this post. Here is an example of what they found in such a blocked-nostril test with one particular mole.
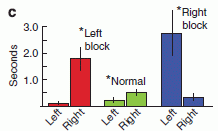
|
In this test, the mole was offered a piece of food, and its path to get to the food was measured. We'll look at exactly what was measured in a moment, but for now simply note that a shorter time -- a lower bar -- is "better". There were three test conditions: normal, and with one or the other nostril blocked.
|
You can see that the results for "normal" were the lowest. With one nostril blocked, the results show one high bar and one low bar. For example... the red bars are for the case where the left nostril is blocked. The result for "left" is about 0.1 seconds; the result for "right" is nearly 2 seconds. Those times are the amount of time the animal spent to that side of the food source. That is, with the left nostril blocked, it spent much of its extended search time to the right of the food -- on the side of the open nostril.
This is Figure 3c from the article.
|
The overall observation is that blocking one nostril causes the mole to veer to the other side -- toward the side of its open nostril. This result suggests that the mole is using the information from the two nostrils separately to determine the location of the food reward.
News stories:
* Moles Smell in Stereo to Find Food, Dodge Predators. (National Geographic, February 5, 2013. Now archived.)
* Evidence Moles Can Smell in Stereo. (Science Daily, February 5, 2013.)
The article, which is freely available: Stereo and serial sniffing guide navigation to an odour source in a mammal. (K C Catania, Nature Communications, 4:1441, February 5, 2013.)
More about moles: The basis of intersexuality in moles (November 28, 2020).
Other posts about smelling include:
* Copper ions in your nose: a key to smelling sulfur compounds (October 10, 2016).
* What does blue light smell like? (July 18, 2010).
* How do you tell if bees are pessimistic? (August 5, 2011).
* Hard seeds or soft seeds? (April 26, 2013).
April 17, 2013
An ancient navigation device?
April 16, 2013
A team of scientists has reported taking a beautiful natural crystal and making a mess of it -- in two weeks. It's an interesting story.
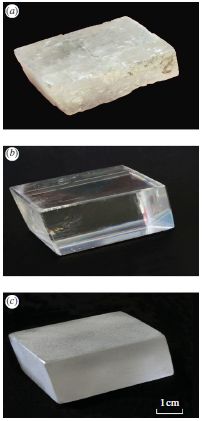
|
Start with the middle picture -- part b. It shows a crystal of calcite, a form of calcium carbonate, CaCO3. It is about 5 cm (2 inches) long; see scale bar at lower right of the entire figure. Beautiful, isn't it?
Part c (bottom) shows this same crystal (from part b) after treatment.
Why is this change of interest? Look at part a (top). This is a photo of a "stone" recovered from a 16th century shipwreck. The scientists think it was originally a nice crystal of calcite (like that of part b), and that it has been degraded by being on the sea bottom for four centuries. The appearance seen in part c is a hint of what prolonged treatment with sea water can do to a crystal of calcite.
Oh, what was that treatment that got us from part b to part c? Sand abrasion, plus two weeks immersion in sea water. As a touch of authenticity, they even used water from the English Channel, the site of the shipwreck.
This is reduced from Figure 1 from the article.
|
It's that old object in part a that is the real story here. It's in bad shape, but it might well have been a single beautiful calcite crystal -- 400 years ago. And if it was, it might have been a navigation device used onboard the ship. Calcite crystals have interesting optical properties; looking through such a crystal allows one to find the sun, even through clouds or when the sun is somewhat below the horizon.
There are references in ancient writings about the Vikings using "sunstones" for navigation. By the time of this ship, magnetic compasses were coming into use, but were still somewhat mysterious. (You thought a compass pointed north? Try telling that to someone at high northern latitudes.) Sunstones may have been still in use, alongside the modern tools. Perhaps this one is an example. If so, it would be the oldest known navigation sunstone recovered from a ship.
News story: First Evidence of Viking-Like 'Sunstone' Found. (M Gannon, Live Science, March 6, 2013.) (This is a replacement news story. The one listed originally is no longer available.)
The article: The sixteenth century Alderney crystal: a calcite as an efficient reference optical compass? (A Le Floch et al, Proc R Soc A 469:20120651, May 8, 2013.)
Here is another news story, about an earlier paper on this work. You may find it useful as background. The Viking Sunstone Revealed? (Science Now, November 1, 2011.)
Musings posts on calcium carbonate include:
* A see-shell story (February 21, 2016).
* Bending a rigid rod (May 17, 2013).
* Quiz: What is it? (March 6, 2012). See the answer.
More about shipwrecks...
* Should physicists be allowed to use lead from ancient Roman shipwrecks? (December 2, 2013).
* A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
* The Antikythera device: a 2000-year-old computer (August 31, 2011).
Infant cured of HIV?
April 15, 2013
It was a big news story in the popular media last month: an infant apparently cured of HIV. It is a potentially important story, so let's look at it. A big caution... It is also an incomplete story. At this point, what we have is a report at a meeting; no scientific paper has been published. If we take the basic facts presented at the meeting as correct, we must remember that this is one case. We think we know what happened, but cannot be sure. The only way to know whether the result holds more broadly is to test it more broadly. It is an exciting enough result that such testing will undoubtedly be done.
Here is the basic story... A child was born to an HIV-infected mother, who had received no treatment. The child was put on HIV-therapy 30 hours after birth -- even before test results were available; the testing showed that the child was indeed positive for the virus. Treatment continued for 18 months, but was stopped. The child has been off therapy for about a year, and appears to be free of HIV. Thus, if we accept the test results showing that the child was indeed infected and is no longer infected, this seems to be a "cure".
What's novel here? Normally, withdrawing treatment of an HIV-infected person leads to a rebound of the virus. The implication is that the virus is "hiding" in the body, in some latent form not susceptible to the ordinary treatment. Treatment works fine, but relax the treatment and the virus rebounds. In the new case, the treatment was relaxed, and no rebound occurred. One possible interpretation is that the treatment began so early after infection that the virus never established its latent or hidden infection. That would be an exciting point, if it really holds.
As we noted at the outset, neither the facts nor the implications are entirely clear. This is a case report, not a controlled experiment. Was the child truly infected? Is it indeed disease-free, or at least virus-free?
If the story is correct as presented, we do not know for sure why it worked here. Perhaps this child was, for some reason, a special case. Nevertheless, we learn from single reports; we at least ask good questions. In this case, the implication is that we should treat immediately after exposure. This might apply to babies born to mothers who were infected but untreated. However, the number of such cases is small in the developed world, and acting on this in the developing world will be challenging.
News stories:
* Toddler 'functionally cured' of HIV infection, NIH-supported investigators report. (NIH, March 4, 2013.) From a funding source.
* Doctors Cure Baby Born With HIV For First Time. (Medical News Today, March 4, 2013.)
* In Medical First, a Baby With H.I.V. Is Deemed Cured. (New York Times, March 3, 2013. Link is now to Internet Archive.) If you read this, try to read it through to the end. Some things are a bit mixed up along the way.
Here is the abstract of the presentation given at the meeting: Functional HIV Cure after Very Early ART of an Infected Infant. (D Persaud et al, 20th Conference on Retroviruses and Opportunistic Infections, March 4, 2013. Now archived.)
More July 21, 2014 (1)...
Here is the article that was later published about that initial report: Absence of Detectable HIV-1 Viremia after Treatment Cessation in an Infant. (D Persaud et al, New England Journal of Medicine 369:1828, November 7, 2013.) Check Google Scholar for an available copy, including an author manuscript at PubMed Central.
More July 21, 2014 (2)...
The follow-up news is not good. Now, a year later, the child is clearly infected.
We had limited information at the time of the initial post, and we have even less at this point on the update. Nevertheless, we should note the setback.
News story: HIV Returns in "Cured" Child -- A Mississippi girl who was thought to have been "functionally cured" of HIV as an infant once again harbors detectable levels of the virus. (The Scientist, July 11, 2014.)
* Previous post on HIV... A simpler assay for detecting low levels of HIV, using gold nanoparticles (January 3, 2013).
* Next... How HIV destroys the immune system (March 3, 2014).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on HIV
Mice with human brain cells
April 13, 2013
Adding human brain cells to mice makes the mice smarter.
That is a true statement about what was reported in a recent article. However, as so often, the attention-getting one-liner -- and indeed the article -- are just small parts of a big story. We want to look at some specifics, but also provide some perspective for the big story. You should go away with more than just the provocative point that mice with human brain cells are smarter.
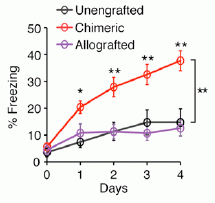
|
Let's start with one of the results. Here is an example of what they found...
Without going into the details for the moment... Three groups of mice were given a behavioral test. You can see that one group, labeled "Chmeric" (red curve), did the best. This is the group of mice with added human brain cells; the other two groups are controls. (The word chimeric refers to the mice being hybrid: part mouse, part human.)
This is Figure 6a from the article.
|
Those results should whet your appetite, so let's look further. What does it mean to say that these mice have added human brain cells?
There are various types of brain cells; the ones added to the mice were astrocytes. Neurons get the most attention, but there is increasing appreciation that astrocytes may be particularly important. One clue is that one of the greatest differences between the brains of man and other animals is in how well human astrocytes are developed. Astrocytes are involved with neural signal transmission, but their role is unclear.
The way the scientists added human astrocytes was to graft human astrocyte precursor cells into the developing brains of newborn mice. Thus the mice were allowed to develop their brains in the presence of the human cells. In fact, the human astrocytes integrated into the mouse brains, but appeared to be the well-developed astrocytes typical of humans. Further, the human astrocytes resulted in improved performance of the mice on certain tests, as illustrated with the graph above.
One of the controls was engrafted using the same procedure but with mouse cells; this control is labeled "Allografted". It is something of a sham control; the animals underwent all the steps of the procedure, but did not get the treatment itself. The other control, "Unengrafted", did not undergo the grafting procedure; that is, these are just normal mice.
Here is one possible interpretation of these results... Astrocytes play a supporting role in the brain, and the extensive development of human astrocytes was an important part of humans developing more advanced brains. When the human astrocytes are in mice, they provide better support there, too, thus enhancing at least some mouse brain functions. It's important to take this as a "possible interpretation", something that can help you see where the results might fit. It is certainly not proven; much more work is needed.
So what's the bigger story? They have an experimental system to study astrocytes. These cells are not well understood. One aspect of the work is that they are able to develop the precursor cells from people with various neurological conditions. They can then add astrocytes reflecting various diseases into the mice, and study the defects experimentally. There is much more to come from this experimental system -- and it is not about making mice smarter.
News stories:
* Using Human Brain Cells to Make Mice Smarter. (Science Daily, March 7, 2013.)
* Using human brain cells to make mice smarter. (Medical Xpress, March 7, 2013.)
* News story accompanying the article: Do Your Glial Cells Make You Clever? (R J M Franklin & T J Bussey, Cell Stem Cell 12:266, March 7, 2013.)
* The article: Forebrain Engraftment by Human Glial Progenitor Cells Enhances Synaptic Plasticity and Learning in Adult Mice. (X Han et al, Cell Stem Cell 12:342, March 7, 2013.)
Video. There is a 5-minute promotional video with two of the senior authors describing the work. It is linked to various stories, and is also available at YouTube video.
Also see...
* As we add human cells to the mouse brain, at what point ... (August 3, 2015).
* A drug that delays neurodegeneration? (June 14, 2013).
* Fish with bigger brains may be smarter, but ... (January 25, 2013).
* The smartest chimpanzee? (September 29, 2012).
* Making smarter flies (July 18, 2012).
* Smart dust: A central nervous system for the earth (July 20, 2010).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
Caffeine boosts memory -- in bees
April 12, 2013
Who would try feeding caffeine to bees? Coffee plants. (And some citrus plants.) The nectar of coffee flowers contains caffeine -- just a little, not enough to taste too bitter. Perhaps enough to make the bees come back.
A new article looks at how bees respond to caffeine. The basic experimental design is familiar. Bees are offered choices. A scent leads to a reward -- of sugar. The question is whether the bees learn to associate the scent with the reward. In this case, one variable was the level of caffeine.
The following graph shows how the bees responded.
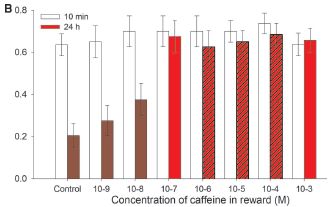
|
The y-axis is the fraction of "correct" responses. The x-axis is the concentration of caffeine (log scale).
The open bars show how the bees respond 10 minutes after learning. They get it right about 60-70% of the time, regardless of the caffeine level.
The colored bars (all of them, regardless of the color markings) are for 24 hours after the learning. At 24 hr, only about 20% of the responses are right for the control -- with no caffeine. As the caffeine level rises, so does the performance of the bees in this learning test.
|
At the higher caffeine levels, the bees do as well at 24 hr after learning as they had done at 10 min. At 24 hr, their "score" here has improved about three-fold by having caffeine.
Three of the red bars are "hatched" (marked with diagonal lines). These three show the levels of caffeine in the natural nectars the scientists examined (from coffee and citrus flowers). The level of caffeine that affects the bees' performance is in the range found in natural nectars.
This is Figure 2B from the article.
|
We see, then, that caffeine can enhance the ability of bees to remember what they learned, and that the level of caffeine required is about what is found in the nectar of some flowers. Thus the scientists suggest that what they studied here, under lab conditions, is likely to be relevant "in the field". They suggest that the plant benefits from providing some caffeine in its nectar, by getting more pollination. It's already known that caffeine at high levels is an insect repellent; finding a beneficial effect of low caffeine would be an interesting development. The results here are consistent with that, but certainly not sufficient. For example... Would it be possible to do field studies and show that increased caffeine leads to more pollination?
News stories:
* Bees Get a Buzz from Flower Nectar Containing Caffeine. (Science Daily, March 7, 2013.)
* Coffee and Citrus Plants Boost Bee Memory With Caffeine. (C Wilcox, Science Sushi (Discover blog), March 7, 2013.)
* News story accompanying the article: Neuroscience: Caffeine Boosts Bees' Memories -- Caffeine in floral nectar enhances the memory of bees for the flowers' scent by altering response properties of neurons in the bee brain. (L Chittka & F Peng, Science 339:1157, March 8, 2013.)
* The article: Caffeine in Floral Nectar Enhances a Pollinator's Memory of Reward. (G A Wright et al, Science 339:1202, March 8, 2013.)
Posts about caffeine or coffee include...
* Added February 25, 2026.
Are coffee stains useful -- in electron microscopy? (February 25, 2026).
* Chocolate: 1200 years old (February 18, 2013).
* Your desire for caffeine: It may be in your genes (May 31, 2011).
More on bees:
* What should a plant do if it hears bees coming? (December 10, 2019).
* Bees and flowers: A 30-volt story (June 21, 2013).
* Novelty-seeking behavior (May 26, 2012).
* The traveling bumblebee problem (January 11, 2011).
More about pollination:
* What if there weren't enough bees to pollinate the crops? (March 27, 2017).
* A "flower" that bites -- and eats -- its pollinator (December 27, 2013).
More about memory: Near-death experiences: are the memories real? (August 11, 2013).
Also see: Why some citrus fruits are so sour (April 22, 2019).
April 10, 2013
SO2 reduces global warming; where does it come from?
April 9, 2013
A major problem in studying "global warming" is that the effects are quite small over short time scales. Not only small, but also variable. A 100-year trend of major warming may include years, or even decades, with little or no warming.
The decade of the 2000s was such a decade, with little change in global temperature. We noted this in an earlier post, which showed that increased sulfur emissions were likely responsible [link at the end]. What was not clear was the major source of the sulfur emissions. Some work suggested that volcanoes were the major source and some suggested that increased burning of coal (especially in Asia) was the major source. Both emit sulfur, in the form of sulfur dioxide, SO2. Atmospheric SO2 leads to aerosols, which cool the planet.
A new article offers some resolution to that issue. The authors show that, for the decade of the 2000s, the sulfur emissions from small to medium-sized volcanoes were the major sulfur source. This is an important finding, since the smaller events were often neglected in earlier work; they may be small, but there are many of them. (The contribution of occasional large volcanic eruptions to cooling has long been recognized.)
The following graph is an example of their findings.
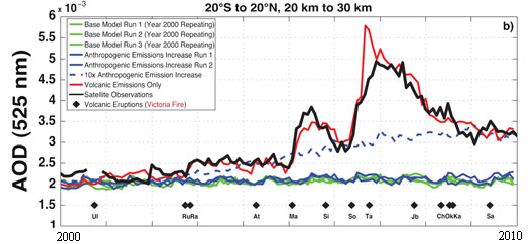
|
The graph shows a measure of atmospheric aerosols over time, for various situations. The y-axis is the aerosol optical depth (AOD), a measure that largely reflects the sulfur emissions in the atmosphere. The x-axis is time, from the year 2000 to 2010. This particular graph is for the equatorial region, between 20° S and N.
There are several curves. Let's look at some of them. The black curve (labeled "Satellite observations") is the observed result: how much aerosol was found by actual measurement. The other curves are all based on modeling done in the new article. The red curve is their model result if they include only "volcanic emissions". The blue curves are their model results if they include only "anthropogenic emissions", that is, human-caused emissions, such as from burning coal. The solid blue curves are for their best estimate of these emissions; the dashed blue curve shows what their model predicts if the anthropogenic emissions were 10-fold higher than their estimates.
This is Figure 1b from the article.
|
You can see that the aerosol levels predicted based on volcanic emissions (red curve) agree rather well with the aerosols observed (black). Aerosols predicted from anthropogenic emissions, even elevated 10-fold (dashed blue curve), do not match the actual record.
A caution... The graph above is for the equatorial region. The full figure in the article also includes north and south temperate regions. The results for those regions are much less clear than for the equatorial regions, although the volcanic contributions seem most important.
The article is a useful contribution, with more advanced modeling. It contributes to our understanding of short-term fluctuations in climate. It shows an important role for emissions from smaller volcanoes.
News story: Volcanic aerosols, not pollutants, tamped down recent Earth warming. (American Geophysical Union, March 1, 2013.)
The article: Recent anthropogenic increases in SO2 from Asia have minimal impact on stratospheric aerosol. (R R Neely et al, Geophysical Research Letters 40:999, March 16, 2013.)
Background post: Why isn't the temperature rising? (September 12, 2011). I do encourage you to go back to this post to fill in the story.
Also see:
* Predicting the "side-effects" of geoengineering? (September 23, 2018).
* Geoengineering: the advantage of putting limestone in the atmosphere (January 20, 2017).
* National contributions to global warming (June 25, 2014).
* When does global warming occur: day or night? (October 28, 2013).
* Climate change and hare color (May 10, 2013).
* Sulfur dioxide in the atmosphere of Venus (February 16, 2013).
Can one rat know what another rat is thinking?
April 8, 2013
Sure. Send the brain signal from one rat to the other.
A new article reports doing just that. We note it briefly...
A recent post showed that a rat could respond to an infrared (IR) sensor that was connected to its brain [link at end]. As a result, the rat gained the ability to respond to IR light, effectively adding a new sense to its sensory repertoire. The new work is from the same group of scientists; in some ways, the new work is similar to that reported in the earlier post. Each is a small step in a big field involving learning about brain signals.
The authors refer to the new work as establishing a brain-to-brain interface (BTBI). The general plan of an experiment is as follows... One rat is given a behavioral test, and makes a decision. Electrodes implanted in the brain of this rat transmit brain signals from this "encoder" rat. Those signals are then transmitted, in real time, to a "decoder" rat, whose actions are then observed. Both rats face the same situation; it turns out that the decoder rat does what the encoder rat had chosen -- with a significant probability.
In one test, just to emphasize the point, the scientists connected two rats on different continents. The encoder rat, making the original decision, was in Brazil; its brain signal was connected to a decoder rat in the United States, who acted on the decision made by the rat in Brazil. How did the signal get from one to the other? Via the Internet, of course. The decoder (receiver) rat did just fine at making use of the information.
This work has something of a "stunt" aspect, and some of the discussion of it includes hype about what might be done in the future. Ok, but the point is that this is all part of the big story of learning about brain signaling. In the IR post, rats learned to use the signal from an external electronic device. In the current post, brain signals are obtained from one rat and transmitted to another. In other work, we have already noted examples of humans benefiting from such applications, even though they are quite primitive at this time. As to developing computers based on such interconnected animals... Well, that illustrates a line of work the authors want to pursue. It's good that they are enthusiastic about their work. Let's see what they learn from it.
News stories:
* Intercontinental mind-meld unites two rats -- But critics are skeptical about predicted organic computer. (Nature News, February 28, 2013.)
* First direct brain-to-brain interface between two animals. (Kurzweil, March 1, 2013.)
The article, which is freely available: A Brain-to-Brain Interface for Real-Time Sharing of Sensorimotor Information. (M Pais-Vieira et al, Scientific Reports 3:1319, February 28, 2013.)
Movies. There are two movie files for this work. One shows a couple of test sequences. The second is an interview with the lab head about the work. You can find them at the lab web page: First brain-to-brain interface allows transmission of tactile and motor information between rats. (Nicolelis lab, Duke University.) (The first is also available with the article, at the journal web site.)
Background post: Can rats touch infrared light? (February 25, 2013). As noted, this work and the current work are from the same lab. This post includes links to other relevant Musings posts.
More...
* The brain-machine interface -- at the World Cup (July 2, 2014).
* Using your brain waves to log on to the computer (April 29, 2013).
And more... Computer scientist thinks; psychologist moves finger (September 24, 2013).
More about enhancing rat brain function... Can blind rats learn to use a geomagnetic compass? (June 29, 2015).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
Thanks to Borislav for alerting me to this work.
Bacteria can make mouse stem cells
April 6, 2013
Briefly noted...
Last year's Nobel Prize for Medicine or Physiology was awarded, in part, to Shinya Yamanaka for discovering how to reprogram adult cells from the mammalian body back to a stem-cell-like state, known as induced pluripotent stem cells (iPSC). Turns out that some bacteria do something very similar. Leprosy bacteria.
Leprosy bacteria infect nerve cells -- a specific type of nerve cell known as Schwann cells. In the new work, scientists found that the bacteria converted some of these nerve cells to a stem-cell-like state; these stem cells could migrate, thus spreading the bacteria. Overall, it seems that the bacteria-induced stem cells promoted both the dispersal of the bacteria, as well as some protection from the host immune system. Further, the loss of the nerve cells probably contributed to the nervous system damage that is a characteristic of leprosy. The work here is in mice; we presume, for now, that the infection follows a similar course in humans, though that has not yet been examined.
That bacteria can do what we can do should not be surprising. The lesson from Yamanaka with iPSC is that a small number of specific proteins can induce such changes in the state of differentiation. Bacteria can change the levels of host proteins; it is not surprising that bacteria can also induce changes in differentiation state. It is not surprising that they can, but it is new to find that they do.
An odd story, perhaps, but also potentially important. The new work shows that bacteria make stem-cell-like cells, but we know little about how they do it at this point. Clearly, further work will be done to learn the details of the process. This is potentially important, in two ways. First, it represents an improved understanding of the leprosy bacteria; perhaps over time it will lead to improved treatment. Second, we want to pursue this to see what the implications might be for us making stem cells.
News story: Bacteria's hidden skill could pave way for stem cell treatments. (Phys.org, January 17, 2013.)
The article: Reprogramming Adult Schwann Cells to Stem Cell-like Cells by Leprosy Bacilli Promotes Dissemination of Infection. (T Masaki et al, Cell 152:51, January 17, 2013.)
Recent post on stem cells: The role of the immune system in making stem cells (February 8, 2013).
I have more on stem cells on my page Biotechnology in the News (BITN) - Cloning and stem cells.
Why the facial tumor of the Tasmanian devil is transmissible: a new clue
April 5, 2013
Briefly noted...
We previously noted the case of the facial tumor of the Tasmanian devil [link at the end]. An unusual feature of Devil facial tumor disease (DFTD) is that it is transmissible. The animals bite each other -- and transmit the cancer; the population of Tasmanian devils has become endangered.
Transmissible cancers are extremely rare, and not understood. Usually, the immune system serves as a barrier to cancer transmission to a new host, even of the same species. Obviously that barrier is not effective with DFTD. A new article offers some explanation of what is going on with this transmissible tumor.
Briefly, the scientists find that the tumor is failing to display tumor antigens, thus making the tumor invisible to a new host. That's different from what was expected, that the devil tumor might not have any distinctive tumor antigens. Further, they think that the failure to display the tumor antigens is not due to a mutation in the display system, but to some operational problem. It's there, but is turned off. In fact, in the lab they can get the tumor to display antigens that could be used to target it.
What are the implications? It's too early to know, but at least it represents a better understanding of a novel disease. Knowing a more specific cause of the problem allows work to proceed focusing on that specific cause. There are tumor antigens and maybe they can be expressed. Further, knowing that poor antigen display is a key issue raises some hope that a vaccine might be useful.
News story: Hope for Threatened Tasmanian Devils. (Science Daily, March 11, 2013.)
The article, which is freely available: Reversible epigenetic down-regulation of MHC molecules by devil facial tumour disease illustrates immune escape by a contagious cancer. (H V Siddle et al, PNAS 110:5103, March 26, 2013.)
Background post: The devil has cancer -- and it is contagious (June 6, 2011). Includes pictures.
An important follow-up: Immunization of devils: a treatment for a transmissible cancer? (April 24, 2017).
Another transmissible cancer? ... Is clam cancer contagious? (April 21, 2015).
More about immune systems: Bach and the immune system (August 26, 2013).
More about epigenetic marks: A DNA test that can distinguish identical twins (July 17, 2015).
April 3, 2013
Propionibacterium acnes bacteria: good strains, bad strains?
April 1, 2013
Propionibacterium acnes (P acnes) bacteria have long been associated with acne. However, the nature of the relationship is not clear. A new article offers some insight. By using more refined analysis, the scientists show that there are various kinds of P acnes bacteria -- with different relationships to the disease.
In the new work, scientists sampled the skin of 101 people, about half of whom had acne. They analyzed the bacteria found on the skin. They determined the prevalence of P acnes, but they also went further and characterized the type of P acnes.
The overall prevalence of P acnes was similar in people who did or did not have acne. However, the results for specific strains were quite different. Here is a brief summary of the results for three strains.
| Strain
| % of isolates,
people with acne
| % of isolates,
people with clear skin
|
| RT1
| 48%
| 52%
|
| RT5
| 99%
| 1%
|
| RT6
| 1%
| 99%
|
As an example of how to read the table... The first row lists the results for a strain called RT1. Of all the isolates of this strain, 48% were found in people with acne, and 52% in people who did not have acne.
|
This is a condensed version of Table 1 from the article.
|
The table lists three strains of P acnes. As noted, strain RT1 is found about equally in people with or without acne. But the other two strains give quite different results. Strain RT5 is found almost entirely in people with acne, and RT6 is found almost entirely in people without acne.
Not all P acnes are equal! Recognizing this may be an important step toward understanding the relationship of these bacteria to the disease. However, we must once again emphasize that we do not know what that relationship is. For example, the results here are consistent with two quite different models...
* It is possible that strain RT6 is a "good" strain, which helps prevent the growth of a "bad" strain such as RT5. In this case, providing people with the good strain might be beneficial.
* However, it is also possible that acne creates conditions on the skin where strains such as RT5 can grow; in this model, the bacteria are a result of the disease, not a cause.
These alternative models are not new. And there are more possibilities. What's new is that we now know it is not sufficient to just say P acnes; we need to look at specific strains. Thus the new article does not solve the acne problem, but it allows a new type of testing to proceed.
News story: Got Pimples? You May Need Better Bacteria. (Science Now, February 27, 2013.)
The article: Propionibacterium acnes Strain Populations in the Human Skin Microbiome Associated with Acne. (S Fitz-Gibbon et al, Journal of Investigative Dermatology 133:2152, September 2013.)
More about the bacteria associated with acne:
* Acne, grapevines, and Frank Zappa (August 1, 2014).
* A virus that could treat acne? (October 21, 2012)
More competition between skin bacteria... Staph fighting Staph: a small clinical trial (April 8, 2017).
A broader view of our microbes: Sharing microbes within the family: kids and dogs (May 14, 2013).
The benefit of providing alcohol to the eggs
March 30, 2013
Given a choice, fruit flies may choose to lay their eggs where there is a high concentration of alcohol (ethanol). Why? To protect their offspring from parasitic wasps, which would lay their eggs in the larval flies. The wasps cannot tolerate the alcohol. On the other hand, the flies, whose natural environment is fermented fruit, can tolerate alcohol.
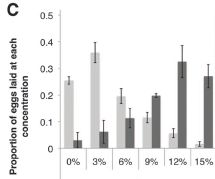
|
This figure shows the basic observation.
Female fruit flies were offered dishes with various concentrations of ethanol, from 0% to 15%. Some were exposed to female parasitic wasps, which could threaten their offspring; some were not.
The bar height shows the fraction of the flies that laid their eggs at the indicated ethanol concentration.
|
The dark bars are for flies exposed to a female wasp. These flies tended to lay their eggs in dishes with high levels of ethanol. About 80% of them chose one of the three highest levels.
In contrast, flies that were not exposed to a female wasp (light bars) tended to lay their eggs in dishes with low levels of ethanol. About 80% of them chose one of the three lowest levels.
This is Figure 1C from the article.
|
Thus it is clear that the flies respond to the presence of the female wasps in a way that benefits the fly offspring. (The flies do not respond to male wasps -- just to the females, the ones that lay eggs in their offspring.) The authors consider this something of an immune response. More specifically, they call it a behavioral immune response, an interesting idea. They also refer to it as medication; the article title uses this terminology.
How do the flies know there is a female wasp around? The scientists do additional experiments in which they manipulate the sensory responses of the flies. Flies with their olfactory (smell) system disrupted respond normally in these tests. However, flies with their visual system disrupted fail to respond to the wasps. Thus the scientists conclude that the flies detect the wasps -- and distinguish male and female -- visually.
News stories:
* Fruit flies defend eggs using alcohol. (Naked Scientists, February 21, 2013.)
* Fruit flies medicate their larvae with alcohol. (Phys.org, February 22, 2013.)
The article: Fruit Flies Medicate Offspring After Seeing Parasites. (B Z Kacsoh et al, Science 339:947, February 22, 2013.)
More about parasitic wasps: Cockroach should be disinfected before eating it (February 12, 2013).
More on fruit flies:
* "Moonwalkers" -- flies that walk backwards (May 28, 2014).
* Making smarter flies (July 18, 2012).
Bat meets spider
March 29, 2013

|
Sometimes, the spider wins.
"Dead bat (Rhinolophus cornutus orii) caught in the web of a female Nephila pilipes on Amami-Oshima Island, Japan (photo by Yasunori Maezono, Kyoto University, Japan; report # 35)." That's from the figure legend; this is Figure 2 part I from the article.
|
A new article is about spiders catching bats. Apparently, not much was known about the topic. That led a scientific team to do an extensive search to see how many incidents they could find. The article is a compilation of all the reports they found: 52 of them.
The article lists the incidents, and has separate tables for the types of spiders and types of bats involved; the tables include the weights of the predator and prey, as best they know them. And the article includes a figure with 12 photos; one of those photos is shown above.
Most of the reported incidents involved a spider catching a bat in its web, a testament to the strength of spider silk. Some involved active predation by large spiders, such as tarantulas.
With only 52 cases found after an extensive search, it would seem that spiders capturing bats is still an uncommon occurrence. But their point is that it is more common than appreciated, and we don't know how often it occurs in natural settings. Findings of giant spider webs across the entrances to bat caves are intriguing; of course, we don't know what is actually caught in those webs.
The article starts with an overview of some unusual feeding habits of spiders; that is worth a look.
News story: Spiders eat bats all the time, scientists reveal -- The capture and killing of small bats by spiders might be more common than previously thought, show recent studies of bat predation by spiders. (Christian Science Monitor, March 18, 2013.)
The article, which is freely available: Bat Predation by Spiders. (M Nyffeler & M Knörnschild, PLoS ONE 8(3):e58120, March 13, 2013.)
More about spiders and spider silk...
* Spider silk: Can you teach an old silkworm new tricks? -- Update (February 11, 2012).
* Tarantulas in the trees (November 11, 2012).
* Spiders in the sky (February 20, 2013).
More about bats...
* Little yellow-shouldered bats -- and the Guatemalan bat flu (March 30, 2012).
* Should you get a rabies vaccination before boarding an airliner? (May 7, 2012).
* Baseball and violins (May 15, 2012).
March 27, 2013
How many moons hath Pluto? Follow-up
March 26, 2013
Original post: How many moons hath Pluto? (July 20, 2012). That post, less than a year ago, presented the newly discovered fifth moon of Pluto. Aside from the fun of discovery, knowing the moons of Pluto is important because we have a spacecraft on its way to Pluto; the success of the New Horizons mission depends on it not hitting the moons. We ended that post by wondering, at least implicitly, how many more, undiscovered, moons Pluto might have.
Here we have a new study with something of an answer to that question. Now, we need to be clear: they did not find anything; they did not even look. Rather, they ran computer simulations of how they think moon formation occurs around Pluto. The following figure summarizes one of their computer results.
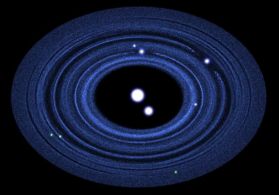
|
Pluto and its moons: what the region might look like, based on computer simulation.
Pluto and its large moon Charon are in the center. The four other known moons are shown in white (above and to the right). Three possible new moons, not yet discovered, are shown in green (near bottom). Also shown, in blue, is the disk of rings of dust from which the smaller moons presumably have condensed.
This is from the Astrobites news story. It is also Figure 13 from the article.
|
That figure shows three more moons. But they don't know how many there might be. One estimate is that there might be ten more -- all too small to detect by current methodologies, but large enough to damage the New Horizons spacecraft. The message? New Horizons will have to fend for itself as it approaches Pluto, looking for unknown moons and adjusting its course as needed.
By the way, that disk around Pluto has never been seen either. It's for New Horizons to find and measure -- data that will feed back to the models of how the moons formed. That is, the computer simulations here and the upcoming observations by New Horizons will not only serve to protect the spacecraft, but also to enhance our understanding of moon formation.
The number of moons of Pluto is both fun and important. Stay tuned.
News story: The Many Moons of Pluto. (Astrobites, March 8, 2013.)
The article: The Formation of Pluto's Low Mass Satellites. (S J Kenyon & B C Bromley, Astronomical Journal 147:8, January 2014.) A copy of the manuscript, as accepted for publication, is freely available at the arXiv.
More about a dwarf planet: Ceres is leaking (February 18, 2014).
A virus with an immune system -- stolen from a host?
March 25, 2013
A virus with an immune system. Fascinating -- and a bit confusing at this point.
As background, we need to introduce the adaptive immune system of bacteria, which was recognized only a few years ago. This system, commonly known as CRISPR, learns from previous infections, and protects the bacteria against future infections by the same agent. It does this by incorporating a bit of the genome of the infecting agent into its own genome, and then making RNA from that copy to watch for and protect against new infections. This immune system is, in some ways, logically similar to our adaptive immune system: it learns (that is, it is adaptive), and it retains its immunological "memory" by changes in its own genome. Of course, the mechanisms of these two adaptive immune systems are quite different.
And now? A virus -- a bacteriophage, a virus that infects bacteria -- that has a CRISPR-type immune system, and uses it to defend against its host.
The story starts rather accidentally. The scientists sequenced the genome of the phage they were studying, and found something that looked like a bacterial CRISPR system. That's odd. Does it function? Here is a test...
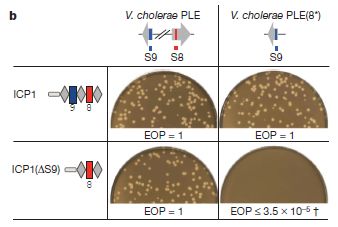
|
This figure shows that the CRISPR "immune system" in the phage is active, and helps the phage grow in its bacterial host.
It's complex, with a lot of jargon. But the key points can be made simply. Let's look. First, look at the results, as shown by the photos. Each photo is showing whether the phage can grow on the bacteria under a specific set of conditions. The little light spots (holes, or "plaques" as they are called with virus work) are a positive result, showing that the phage grew. (The results are also summarized by a number shown immediately below each photo. The number is called efficiency of plating, or EOP. EOP = 1 means the phage grew well; a low EOP means it did not.)
|
You can see that the phage grew well in three of the four cases (lots of plaques, and EOP = 1), and very poorly in the fourth case (no plaques, low EOP). What's special about that case? They had modified the phage and the bacteria so that the phage CRISPR no longer targeted the host. With the phage no longer protected by its "immune system", it was unable to grow.
You can skip down to below the figure explanation if you want; the key idea is above. But, if you'd like a bit more detail...
The experiment shown above involved two bacterial host strains and two phage strains. Two strains of the host bacteria are listed across the top. One (PLE, the wild type) contains two sequences that are relevant here: S8 and S9 (red and blue, respectively). These sequences are part of a host system that helps protect the bacteria against the phage. The bacteria on the right [PLE(8*)] are modified, so that S8 looks different; it still functions, but its specific gene sequence is different. (This involves making so-called silent or synonymous changes, which affect the gene, but not the protein it codes for.)
Two phage strains are shown at the left. The first (ICP1) is wild type; its immune system targets both regions 8 & 9. But the second phage [ICP1(ΔS9)] has been constructed with the response against 9 deleted. The second phage can target 8 but not 9.
Look at the upper left case: wild type phage growing in wild type bacteria. The phage can defend against both 8 and 9 -- and grows fine. This is the key control. Now look at the lower right case. The phage cannot target 8, because 8 has been modified in the bacteria. And the phage cannot target 9 because its response to 9 has been deleted. Thus the phage in the lower right case cannot target either 8 or 9 -- and cannot grow. This shows that its immune response is needed for growth.
(The other two cases involve partial responses. The results here are not easily predicted, and are not part of the key story.)
This is Figure 3b from the article.
|
Overall, they make a good case that the CRISPR immune system is functioning in this virus, and is defending the virus against the host. It is a novel finding.
How did this virus acquire this "immune system"? We don't know. Given what we know about CRISPR, it seems plausible that this bacterial virus "stole" it from a bacterial host. There is a catch, however. The virus grows on Vibrio cholerae bacteria. To our knowledge, this species of bacteria does not contain a CRISPR system. Perhaps there are strains that do carry it, and we do not know about them yet. Or perhaps there is a more complex story somewhere back in history. In any case, this may well be an example of horizontal gene transfer. Perhaps one reason CRISPR is widespread among the bacteria is that viruses are helping to move it around. Are there other viruses with CRISPR systems? How is the CRISPR system maintained in the virus? More mysteries. The current paper is the first report of a phage with what we thought was a bacterial immune system.
News story: Viruses can have immune systems, new research shows. (Phys.org, February 27, 2013.) Too much hype, but it is also a useful overview. (The hype comes from the original news release from the university, and was repeated in many news stories. As an example, the item says that "The study lends credence to the controversial idea that viruses are living creatures...". It does no such thing. The study shows that a virus may have this particular function; it says nothing about the grand status of viruses.)
* News story accompanying the article: Virology: Phages hijack a host's defence. (M Villion & S Moineau, Nature 494:433, February 28, 2013.)
* The article: A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. (K D Seed et al, Nature 494:489, February 28, 2013.)
More on horizontal gene transfer (HGT):
* An extremist alga -- and how it got that way (May 3, 2013).
* GEBA: B -- revisited
or
Horizontal gene transfer: the web of life? a challenge to evolutionary theory?
(March 26, 2010).
More about CRISPR:
* CRISPR: an overview (February 15, 2015).
* CRISPR: What's it doing to help bacteria carry out infections? (September 8, 2013).
* Exploiting the bacterial immune system as a tool for genetic engineering: The Caribou approach (May 4, 2013).
More about cholera bacteria: Designing a probiotic that fights cholera (December 13, 2010).
Bacteria on human teeth -- through the ages
March 24, 2013
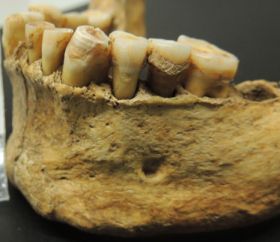
|
A human jaw, labeled as "prehistoric". With teeth. With tooth decay.
This is trimmed and reduced from a figure in the National Geographic news story.
|
What makes this interesting is that scientists were able to extract DNA from the decayed teeth. The calcified dental plaque seems to protect the bacterial DNA from being lost over time. They then identified the types of bacteria that were present. They did this for 34 early European skeletons, spanning several thousand years and various lifestyles. From their results, they concluded that the nature of the human oral microbiota -- the bacteria in our mouth -- changed at two key times in human history. One was the transition from hunter-gatherer to farmer, and the other was the industrial revolution. They further suggest that these changes were important for our understanding of modern oral disease.
It's interesting that they were able to do this. It is another testament to the revolution in sequencing DNA -- and in carefully handling ancient DNA. I think it's fair that the conclusions here should be taken as preliminary. In the big scheme of things, they have a small data set at this point, only 34 individuals. What is important is they have established the approach; more data -- a wider variety of samples -- is bound to come. We are just beginning a story of how the human oral microbiota has adapted to changing human lifestyles.
News stories:
* Calcified Bacteria Sheds Light on the Health Consequences of the Evolving Diet. (SciTech Daily, February 18, 2013.)
* Prehistoric Plaque and the Gentrification of Europe's Mouth. (E Yong, Not Exactly Rocket Science (National Geographic blog), February 17, 2013.) Now archived.
The article: Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. (C J Adler et al, Nature Genetics 45:450, April 2013.)
A recent post on the gut microbiota: Malnutrition: is more (or better) food the answer? (March 8, 2013).
A recent post on the skin microbiota: A virus that could treat acne? (October 21, 2012)
For a broader perspective on our microbiota: Sharing microbes within the family: kids and dogs (May 14, 2013).
More from dental plaque: Division by 14: a new mode of bacterial growth (November 13, 2024).
More from the analysis of old DNA: Tracking the pathogen of the Irish potato blight (June 25, 2013).
More from the study of old teeth:
* How to eat if your jaw looks like a circular saw -- a follow-up (March 8, 2015).
* The case of the missing incisors: what does it mean? (September 13, 2013).
More about tooth decay:
* How the teeth are sensitive to cold (June 14, 2021).
* Is fluoride neurotoxic to the human fetus? (December 13, 2017).
Also see: It's a dog-eat-starch world (April 23, 2013).
There is more about genomes on my page Biotechnology in the News (BITN) - DNA and the genome. It includes an extensive list of Musings posts in the broad areas of genomes and sequencing.
Global warming trend? Independent evidence
March 22, 2013
It's "common knowledge" that there has been a trend of global warming since about 1900. However, this common knowledge is not entirely free of controversy. It is based on ordinary thermometers -- measurements of local temperature at numerous measuring stations around the world. Some aspects of these thermometer measurements have been questioned. For example, these thermometers are sometimes moved, and some may be in urban areas, with increasing local heating from human activity.
Is it possible to get independent evidence on global temperature during this same time period? A new article says yes, and reports that the new evidence is in good agreement with the commonly reported temperature trends.
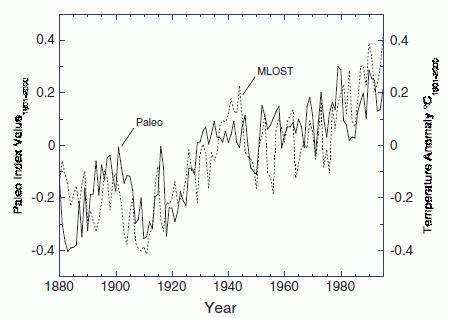
|
This figure summarizes the findings.
There are two curves, each plotted against time, from 1880 to 1995.
The big picture is that the two curves broadly show the same general trend.
|
The dashed curve (axis labeled to the right) is for MLOST, a commonly used measure of temperature trends. (MLOST? It stands for merged land-ocean surface temperature.)
The solid curve (axis labeled to the left) is for the Paleo Index, developed in this new article.
What is this Paleo Index? It's based on measuring other things that the scientists know are "proxy" for temperature. As they say in the abstract, "We compiled the Paleo Index (PI) from 173 temperature-sensitive proxy time series (corals, ice cores, speleothems, lake and ocean sediments, historical documents)." Many of these methods are commonly used in paleontology for determining temperatures for ancient times; the scientists have adapted them to the recent period where they serve as a cross check on the common thermometer records. Importantly, they do not claim that any of these tell them specific temperatures; what they look for is the trend for any particular type of proxy measurement. They then combine all the trend information they get from these 173 proxies into the Paleo Index.
This is Figure 1, top frame, from the article.
|
That is, they have two types of temperature records. One is the traditional modern records, based on thermometers. The other is a measure of temperature without using ordinary thermometers. The two records generally agree. These are independent types of measurements. In general, the criticisms that have been offered regarding the thermometer measurements have no relevance to the Paleo Index.
In comparing the two curves, emphasize the broad pattern. It is not important whether they agree in every detail. (Of course, over time, people may analyze some of the details, and learn from them.) They agree overall, and they agree in some of the details. Two independent types of measurements support the general warming trend since 1880.
News story: Independent Evidence Confirms Global Warming in Instrument Record. (US National Climatic Data Center, NOAA. Now archived)
The article: Global warming in an independent record of the past 130 years. (D M Anderson et al, Geophysical Research Letters 40:189, January 16, 2013.) The paper is a readable overview of the work, with summary graphs such as the one included above. The list of specific methods and the data are in supplements, available at the journal web site. They are very difficult to read!
Video. There is a short promotional video. It doesn't have much depth, but offers a reasonable summary -- and includes some nice pictures. It is available at NOAA: An Independent Record -- Measuring climate change without thermometers. (YouTube, 3 minutes.)
For more about global warming...
* Economic analysis of the damages (and benefits) from climate change (August 26, 2017).
* National contributions to global warming (June 25, 2014).
* When does global warming occur: day or night? (October 28, 2013).
* Climate change: Should we focus on methane? (March 24, 2012).
Do animal bones have something like annual growth rings? (August 7, 2012). This post is about growth rings -- such as the tree rings, or analogous rings in animals. This might be a good example of a paleo proxy: a measurement that reflects temperature, but does not use thermometers. Interestingly, they choose not to use tree rings in this paper. (I am not entirely sure why; it may be a problem of access to the raw data.) Nevertheless, in thinking about the new work in this post, tree ring analysis is a proper example.
More about thermometers: Where is the hottest part of a living cell? (September 23, 2013).
March 20, 2013
The Obama lizard
March 20, 2013

|
Fossil remains of an Obamadon gracilis. It shows some of the teeth, which are the basis of the species name.
This is Figure 1B from the article.
|
About 65 million years ago the dinosaurs became extinct. The common view is that they were the victim of a major calamity, including the collision of an asteroid with Earth. It wasn't just the dinosaurs; a good fraction of species became extinct.
A new article examines the effect of this mass extinction event on snakes and lizards. These groups of reptiles had been thought to survive the extinction rather well. However, the new analysis suggests otherwise.
During the course of the work, the scientists discovered three (extinct) lizard species that had not been previously identified. They named one of them after the US President; it is shown above.
News story: Asteroid That Killed the Dinosaurs Also Wiped out the 'Obamadon'. (Science Daily, December 10, 2012.)
The article: Mass extinction of lizards and snakes at the Cretaceous-Paleogene boundary. (N R Longrich et al, PNAS 109: 21396, December 26, 2012.)
More about extinctions: The 6th mass extinction? (April 4, 2011). The 5th of those mass extinctions is the one discussed in the current post.
More about Obama: Quiz: Barack Obama and polar bears (July 20, 2011).
and then... The Trump moth (January 31, 2017).
More about dinosaurs:
* How the birds survived the extinction of the dinosaurs (June 6, 2014).
* The oldest dinosaur embryos, with evidence for rapid growth (May 7, 2013).
* Do animal bones have something like annual growth rings? (August 7, 2012).
More about lizards: An advanced placenta -- in Trachylepis ivensi (October 18, 2011).
Venus: an unusual view
March 18, 2013
Yes, those are the rings of Saturn. But the post is about Venus. Do you find Venus in there?
Look for a tiny light dot; it is in the upper right. (It's just above one of the darker rings.) That's Venus -- photographed through the rings of Saturn. The Cassini spacecraft just happened to be in the right place at the right time.
If you have trouble finding Venus in the figure above, here is another figure. It includes only a small area, but is blown up. Venus is very near the upper left corner. Venus: focused view [link opens in new window]. In any case, I have noticed that finding the tiny Venus depends on the viewing angle. If you have trouble, move your head, to get a better angle on the image.
Announcement and figure source: Earth's Twin Seen From Saturn. (NASA, March 4, 2013.) The page links to larger versions of the figure.
More about Venus: Sulfur dioxide in the atmosphere of Venus (February 16, 2013).
More from Cassini:
* Quiz: what is it? (April 5, 2017).
* Titan: tides, and the possibility of a sub-surface water ocean (August 4, 2012).
More about the rings of Saturn:
* Briefly noted... Chrysalis and the rings of Saturn? (November 1, 2022).
* The Lord has a new ring (October 12, 2009).
More about rings in the Solar System: Rings for Chariklo (May 9, 2014).
More rings: The largest -- and most distant -- planetary ring system (February 9, 2015).
The proton -- and a 40 attometer mystery
March 17, 2013
How big is a proton? It has actually been measured. Various methods have been used over the years, and an official value for the size of the proton is recognized. Now we have a new measurement -- actually a refinement and confirmation of one reported a few years back. The new measurement has created a new problem.
The new measurement was done by measuring the spectrum of muonic hydrogen. A muonic hydrogen atom is like a hydrogen atom -- but with a muon (instead of an electron) orbiting the single proton. The muon has the same charge as the electron, but is about 200 times heavier; the basic ideas of the structure and properties of an atom should hold for muonic hydrogen, but the numbers are different. Thus, measurement of the exotic muonic hydrogen offers a new approach to measuring the properties of the proton.
We might write an ordinary hydrogen atom as e-p+, where e- is an electron with charge -1 and p+ is a proton with charge +1. Then, a muonic hydrogen atom is µ-p+; µ- is a muon with charge -1.
The scientists measure the spectrum of muonic hydrogen: what wavelengths of light does it absorb? That measures the difference between certain energy levels in the atom, values that are sensitive to the positive charge nearby. Because the muon is much heavier than the electron, it is much closer to the nucleus, and more sensitive to the precise size of the proton. It's all very logical -- just incredibly demanding technically. Muonic hydrogen atoms don't last very long; the scientists have only a few microseconds to take their measurements for each muonic hydrogen atom they produce.
The following figure summarizes the measurements -- old and new -- of the size of the proton. (By "size of proton" here we mean a specific property, called the root mean square charge radius. It is, in simple terms, the size as measured by looking at the charge distribution in the proton.)
|
Look at the line labeled "CODATA (2010)". That line shows the officially recognized value, which is about 0.88 femtometer (1 fm = 10-15 meter). The measurements that were considered in establishing this official value are shown above the CODATA line.
At the bottom there are two (short!) lines labeled "Muonic hydrogen spectroscopy". You can see that they are quite distinct from the CODATA value (and the data behind that value). They are both about 0.84 fm, with quite small error bars.
|
The first of these two muonic hydrogen results was published three years ago, and caused concern. Now, the scientists have carried out further measurements on the muonic hydrogen, as shown by the second value. It agrees with the first. In fact, it is even slightly smaller, and has a smaller error bar.
This Figure is from the news story in Science.
|
Thus we have a discrepancy. At this point, no one understands why. Is there some problem with one set of measurements -- a problem not taken into account in the error analyses so far? Is there some reason why a proton might actually have a different size when combined with a muon? All we can do now is to note the discrepancy and ask some questions. The paper itself ends with an extensive discussion of some of the possibilities. For now, we do not know the size of the proton (to within about 40 attometer). That means that our understanding of this particle, key to atoms and chemistry, is incomplete.
News story: Physicists confirm surprisingly small proton radius. (Phys.org, January 24, 2013.)
* News story accompanying the article: Physics: How Big Is the Proton? (H S Margolis, Science 339:405, January 25, 2013.)
* The article: Proton Structure from the Measurement of 2S-2P Transition Frequencies of Muonic Hydrogen. (A Antognini et al, Science 339:417, January 25, 2013.)
Those who have some chemistry might make a quick estimate of the size of the proton... Atoms are about 0.1 nanometers (1 Angstrom) across, and the nucleus is about 1/100,000 of the size of the atom. Do the arithmetic, and you will get an estimate for the size of a nucleus: about 1 femtometer. Of course, the smallest nucleus is simply a proton. That estimated value is indeed close to the values measured, as discussed above.
More from the atomic nucleus...
* Data on 31 isotopes of cadmium support a new model for the structure of atomic nuclei (December 1, 2018).
* Discovery of the neutron: 80th anniversary (February 27, 2012).
* Quark soup (August 15, 2011).
My page of Introductory Chemistry Internet resources includes a section on Nuclei; Isotopes; Atomic weights. It includes a list of related Musings posts.
Also see... What is the charge on atoms of anti-hydrogen? (July 15, 2014).
More muons... Using your smartphone to detect cosmic rays (April 7, 2015).
Thanks to Greg for helpful discussions of this post.
Carnivorous plants: A blue glow
March 16, 2013
How do carnivorous plants attract prey? Here is one way...

|
The pitcher plant Nepenthes khasiana. It was photographed under UV light; the key feature is the blue fluorescent glow that is induced by the UV light. Importantly, the glow is right at the rim of the pitcher.
This is trimmed from a figure in the National Geographic news story. Pictures of several carnivorous plants photographed the same way are in the article.
|
The glow is barely visible to humans, but is likely to be visible to small animals with good vision in the far blue, especially in dim light. That's the hypothesis. As a test, they coated the fluorescent region to mask the glow; such non-glowing pitchers were less effective at capturing insects. Thus they have at least some experimental evidence for the importance of the glow. Nevertheless, the importance of this phenomenon under natural conditions remains to be investigated.
News stories:
* Carnivorous plant species glow blue to lure prey. (BBC, February 19, 2013. Now archived.)
* Carnivorous Plants Glow to Attract Prey. (National Geographic, February 25, 2013. Now archived.)
The article: Fluorescent prey traps in carnivorous plants. (R Kurup et al, Plant Biology 15:611, May 2013.) This is a nice example of research from a botanical garden, the Jawaharlal Nehru Tropical Botanic Garden and Research Institute in Thiruvananthapuram, India.
More Nepenthes: Underground pitchers (September 6, 2022).
Other posts about carnivorous plants include...
* Look who's dining on baby salamander (November 3, 2019).
* Venus flytrap: converting defense into offense (July 27, 2016).
* Why would a plant have leaves underground? (January 21, 2012).
* How fast can a plant eat? (March 23, 2011).
And also... Carnivorous algae -- that hunt large animals (October 7, 2012).
UV vision:
* Why might it be good to put lights on fish nets? (September 9, 2013).
* Butterflies and UV vision (June 29, 2010). This post discusses an example of an insect with vision in the UV region. (I should note that it is not entirely clear whether the glow of the pitcher plant is in the UV, or merely in the far blue.)
More about vision: What if there was a gorilla in the X-rays of your lungs? (July 26, 2013).
Also see: It's a dog-eat-starch world (April 23, 2013).
Also see: A "flower" that bites -- and eats -- its pollinator (December 27, 2013).
A novel type of polymer -- and its possible relevance to the origin of life
March 15, 2013
The three polymers that carry sequence information in our modern biological world are DNA, RNA, and protein. Among these, it seems likely that RNA is the oldest. Thus we can imagine some primitive stage in which RNA alone -- of those three -- was present, carrying out the roles of all three modern polymers. That stage is sometimes termed the RNA World.
So where did RNA come from? RNA is meaningful only in the polymer form, and it is not obvious how polymeric RNA might have formed originally. A new article offers a hint of how this might have happened. The scientists make small chemical subunits that have some features of RNA bases; importantly, they spontaneously "pair" and assemble into a polymer in aqueous solution.
|
The figure gives the idea. A good place to start is perhaps the middle. This shows a cartoon structure of one level in the polymer. It is something like one base pair in RNA (although you will notice it is actually a triplet, not a pair). When they put these pieces into water, they stack, forming a polymer; this is shown at the right.
The left frame shows the details of the chemical structures. Note that the subunits, TAPAS and CA, can hydrogen bond to each other, similarly to how bases in RNA (or DNA) hydrogen bond to each other.
This is from the Science Now news story; it is also probably the same as Figure 1A from the article.
|
That stacking is the key point. They show that it is possible for something like an RNA base pair to spontaneously stack into a polymeric form. And they understand why the stacking occurs: it is due to the hydrophobic interaction between the single units -- just as with modern nucleic acids.
How close is this to being RNA? We have already noted that they formed a 3-part structure, not a 2-part structure as in the modern base pair. But the individual parts are somewhat like the RNA bases, in that they include nitrogen-containing aromatic rings with hydrogen-bonding groups on the edges. In fact, one of the subunits has a pyrimidine ring, which is the core of two of the modern nucleic acid bases. Further, the polymers they get are quite long, as much as 18,000 units long -- long enough to reasonably call them gene-sized.
Thus they suggest that that they have revealed a possible pathway for how RNA-type polymers might have originally formed. Is this what really happened? We have no way to know, but it is interesting chemistry to demonstrate possibilities.
News stories:
* Molecules Assemble in Water, Hint at Origins of Life. (Science Daily, February 20, 2013.)
* Self-Assembling Molecules Offer New Clues on Life's Possible Origin. (Science Now, February 11, 2013.)
The article: Efficient Self-Assembly in Water of Long Noncovalent Polymers by Nucleobase Analogues. (B J Cafferty et al, Journal of the American Chemical Society (JACS) 135:2447, February 20, 2013.)
Other posts that may relate to the origin of RNA or, more broadly, to the origin of life include...
* An RNA replicator network (March 26, 2022).
* Another possibility for the origin of ribo- and deoxyribo- nucleosides (July 5, 2020).
* The magnesium dilemma: a step toward understanding how RNA might have been made in "protocells" (February 22, 2014).
* The origin of reactive phosphorus on Earth? (July 5, 2013).
* Did life start in a geothermal pond? (February 28, 2012).
* On the road to life? (May 18, 2009).
A good book on the origin of life is noted on my page Books: Suggestions for general science reading: Deamer, First Life (2011).
A post on the history of the Central Dogma, which ties together the roles of DNA, RNA and protein: Central Dogma of Molecular Biology (August 16, 2011).
There is a connection of this story to an unfortunate story of child poisoning a few years back. That story involved milk that had been contaminated with melamine. Investigation showed that the toxicity was due to a complex formed between melamine and cyanuric acid. The latter is the CA of the current work; melamine is very similar to the blue part of TAPAS. For more on that melamine story, see my page of Internet resources for Introduction to Organic and Biochemistry, in the section on Amines, amides.
Melamine toxicity: possible role of gut microbiota (April 21, 2013). More about that story of melamine toxicity.
More about self-assembly: Nanolympiadane: chemical synthesis of the Olympic rings (October 11, 2020).
March 13, 2013
Fair skin and cancer: What is the connection?
March 12, 2013
It's well known that redheads -- people with reddish hair and fair skin -- get more melanoma, a serious form of skin cancer. Why? The common view is that, with their light skin, they are more sensitive to ultraviolet (UV) radiation from the sun. A new article suggests that may not be the whole story.
In the new work, scientists developed a strain of mice that corresponds to such redheaded (or fair-skinned) people. These mice developed more skin cancer even when protected from UV. Therefore, it seems likely that the mutation leading to redheadedness causes an intrinsic susceptibility to cancer.
The following figure gives an idea of what they did.
|
Part a (top) shows the three color types of mice they used. From left, they are black, red, and albino; the names are chosen to emphasize the relatedness to human hair and skin types. In fact, both the red and albino mice carried mutations like those found in humans with those conditions.
All strains used here carried a mutation that increased the chances of melanoma.
Part c (bottom) gives an example of survival curves found with these strains of mice. The conditions involve no UV exposure, or other outside source of DNA damage.
You can see that the red mice (red curve) show the poorest survival.
This is Figure 1 parts a and c from the article.
|
The basic observation, then, is that mice with a gene for redheadedness show poorer survival than other mice -- even when protected from the sun (or other external sources of DNA damage). Whether this holds for humans is not known. The interpretation is that the gene for redheadedness somehow generates DNA damaging agents within the animal. They suggest that this has something to do with the oxidative nature of pigment synthesis; it's interesting that blocking pigment synthesis, with the albino gene, protects the mice.
Caution... Even if this is all correct and relevant for humans... This work does not mean that redheads can freely partake of the sunshine. It actually does not provide any evidence on that point, but it is likely that melanoma in redheads is enhanced by the UV from sunshine. What's new here is to suggest that protecting against the UV does not completely solve the problem. That is, redheads may be at risk for two reasons: susceptibility to UV from sunshine but also by making DNA damaging agents internally.
News story: Unexpected factor contributes to melanoma risk in red-haired, fair-skinned individuals -- Red/blond pigment may actively participate in melanoma formation, identifying possible new prevention strategy. (Massachusetts General Hospital, October 31, 2012. Now archived.) (This is a replacement news story; the one originally listed is no longer available.)
The article: An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. (D Mitra et al, Nature 491:449, November 15, 2012.)
Also see:
* Anti-oxidants and cancer? (October 18, 2015).
* A possible hazard of using compact fluorescent light bulbs (November 13, 2012). This post deals with possible leakage of UV light from CFLs.
* Butterflies and UV vision (June 29, 2010).
* A gene for breast cancer: what does it do? (May 4, 2010). This is about another gene that causes cancer. It includes some discussion of how UV causes cancer or mutations.
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Cancer.
What caused the C-14 increase in the year 775 AD?
March 11, 2013
In an earlier post, we noted a recent report of an unusually high content of carbon-14 (14C or C-14) in tree rings for the year 775 AD [link at the end]. Since C-14 production is stimulated by gamma (γ) rays, the tree ring anomaly pointed to an anomalous source of gamma rays for that time. Some possible sources were considered; none fit with what was known.
A new article suggests another possible source: a short gamma ray burst (GRB). Such a GRB, lasting less than two seconds, might have been due to a collision of two "small" objects, such as neutron stars or white dwarfs. It is plausible that such an event, at a suitable distance, might not have been observed.
It's an interesting story. But remember, the scientists have no evidence whatsoever for such an event. What they do is to add another type of event to the list of candidates, and explain why it is perhaps more reasonable than the others suggested so far. In fact, the paper is probably a more thorough discussion of the candidates than we have seen so far. They do suggest that it might be possible for astronomers to find the object that gave rise to the gamma ray burst.
This remains a story in progress. It started with careful observations of tree rings, with one-year resolution. Those observations of tree rings are leading to questions about the nature of our galactic environment.
News story: Did an 8th Century Gamma Ray Burst Irradiate Earth? (Science Daily, January 21, 2013.) For the most part, this is a good overview. (However the part about beryllium-10 is partly incorrect; ignore it.)
The article, which is freely available: A Galactic short gamma-ray burst as cause for the 14C peak in AD 774/5. (V V Hambaryan & R. Neuhäuser, Monthly Notices of the Royal Astronomical Society 430:32, March 21, 2013.) In some ways, this is a quite readable paper, as it summarizes the main arguments. However, be forewarned that you will be overwhelmed with numbers; with a little judgment, you can read past most of them and get the main ideas just fine.
Background post: Tree rings, carbon-14, cosmic rays, and a red crucifix (July 16, 2012). The observed C-14 blip was between the years 774 and 775; for simplicity, I have referred to this as 775.
Helicoprion -- a fish with 117 teeth, arranged in a spiral
March 9, 2013

|
A fossil of Helicoprion. The structure is 15-20 centimeters across.
The name means "spiral saw".
From Wikipedia: Helicoprion.
|
Helicoprion has long fascinated biologists. It has been extinct for 250 million years. It doesn't fossilize very well -- because it doesn't have bones; it is a cartilaginous fish (think shark, perhaps, though this one is not a shark). The prominent fossil structure -- as you can see -- is a whorl of teeth: 117 of them in the case studied below, in 3 1/4 turns. A whorl of teeth. Where do they go in the animal, and what do they do? Even deciding that these really are teeth and are in the mouth is not simple, given the sparse evidence.
A new article reports new analysis of a Helicoprion fossil. The scientists did computer tomography (CT) scans of the fossil; this provided information beyond what is visible from the surface. The specimen they examined includes some bits of cartilage, including some pieces of skull. The CT evidence allowed them to conclude that the tooth whorl was attached to the back of the lower jaw.
And that leads to...

|
A drawing of what this fish might have looked like, incorporating the newest evidence.
The entire fish may be 3-4 meters long.
This is Figure 1 part l ("el") from the article.
|
The suggestion is that the animal makes teeth throughout life. The newest and largest teeth are on the outside. The older, smaller teeth are wrapped into the inner part of the whorl. There are, it seems, no teeth in the upper jaw. Exactly what the animal does with this whorl of teeth is not clear. The teeth may help snare prey. Further, as the jaw closes, the teeth whorl may help carve the meal. A guess, based on limited structural information.
The article and the news stories about it contain many pictures. Some are photos of fossils. Some are drawings of what the fish might have looked like, with various views over many decades. Again, any attempt to draw the fish is just a "best guess" based on information and ideas available at that time. For the most part, the only real evidence was a whorl of teeth. But the pictures are fun.
News stories:
* Paleontologists Solve Mysteries of Permian Whorl-Toothed Shark. (Sci.News, February 28, 2013.)
* Buzzsaw Jaw Helicoprion Was a Freaky Ratfish. (B Switek, National Geographic Blog, February 26, 2013.)
The article: Jaws for a spiral-tooth whorl: CT images reveal novel adaptation and phylogeny in fossil Helicoprion. (L Tapanila et al, Biology Letters 9:20130057, April 23, 2013.)
Follow-up: How to eat if your jaw looks like a circular saw -- a follow-up (March 8, 2015).
More on fish:
* Microraptor was piscivorous (May 25, 2013).
* Fish with bigger brains may be smarter, but ... (January 25, 2013).
* Did you see what the sawfish sawed? (April 27, 2012). There is no particular relationship between the sawfish and Helicoprion.
More on teeth:
* How the teeth are sensitive to cold (June 14, 2021).
* The "hobbits": dentition suggests they were a distinct, dwarfed human species (November 30, 2015).
* A rodent that can't chew (November 5, 2012).
* Analysis of teeth confirms that Regourdou was right-handed (September 7, 2012).
More on cartilage: The role of zinc in arthritis (July 18, 2014).
Malnutrition: is more (or better) food the answer?
March 8, 2013
Perhaps not, according to a new article, which suggests that the underlying problem in some malnourished children may be the microbes in the gut. It is a clever piece of work, and it opens up new approaches to considering malnutrition.
Here is the idea. The scientists study the specific type of malnutrition known as kwashiorkor. It's a disease where a distinctive feature is the swollen belly; you've probably seen pictures. They focus on discordant twins -- twins where one has the disease and one does not. It is assumed, reasonably enough, that the twins had more or less the same diet. The key finding was that those with and those without the disease had different gut bacteria. That is interesting, but alone tells us little. In particular, it says nothing about any causal relationship. Does the disease cause a change in the gut microbiota, or is it possible that the gut microbiota changes for some reason, and that contributes to the disease process?
As a second test, they fed children with the disease a "therapeutic diet" -- an inexpensive nourishing food product used to treat malnutrition. A simple summary of what they found is that there was some improvement in the gut microbiota, but it was incomplete.
A third type of experiment is perhaps the most intriguing. They tested the effect of the various collections of microbes on nutrition. Of course, they did this in mice, not humans. The basic finding was that mice with gut microbes from the healthy kids fared better than the mice with gut microbes from the kwashiorkor kids. This would appear to be a direct demonstration that the gut microbes of the kwashiorkor kids are themselves part of the problem. This is a well-controlled experiment, in mice. We do not know its relevance to humans; that has not even been tested.
Let's, for the sake of discussion, assume that the mice result holds with humans. It raises numerous questions -- some of which are probably easy to address. Is the disease onset due to some event that affects the gut microbiota, and makes the kids unable to metabolize their food properly? Could we diagnose children likely to get kwashiorkor before they get it, by testing their gut microbes? If so, could we do something about it? Does knowing about the role of microbes lead to a treatment? Would replacement of gut microbes help? Or would a different choice of foods help, by helping to stimulate the preferred microbes? Lots of question, as so often the case when we get a novel intriguing result. It's a clue, a piece of the puzzle.
News stories:
* Gut microbes at root of severe malnutrition in kids. (Washington University, St. Louis, January 30, 2013.) News release from the lead institution.
* Gut Microbes Contribute to Mysterious Malnutrition. (E Yong, Not Exactly Rocket Science (National Geographic blog), January 30, 2013.)
* News story accompanying the article: Microbiology: Undernutrition - Looking Within for Answers. (D A Relman, Science 339:530, February 1, 2013.)
* The article: Gut Microbiomes of Malawian Twin Pairs Discordant for Kwashiorkor. (M I Smith et al, Science 339:548, February 1, 2013.)
Among posts about the microbiota...
* Treating malnutrition by improving the gut microbiome (May 29, 2021). More, from the same lab. (But it is not a direct follow-up of the same situation.)
* Our microbiome: a caution (August 26, 2014). The hype of microbiome research.
* Melamine toxicity: possible role of gut microbiota (April 21, 2013). Was the toxicity of melamine in the poisoning incidents a few years ago mediated by gut bacteria?
* Bacteria on human teeth -- through the ages (March 24, 2013). The oral microbiota.
* A bacterial cocktail to fight Clostridium difficile (January 19, 2013). An example of treating a disease by directly changing the gut microbiota -- again in mice.
* Antibiotics and obesity: Is there a causal connection? (October 15, 2012). Does the gut microbiota affect obesity -- as well as malnutrition?
More about twins: A DNA test that can distinguish identical twins (July 17, 2015).
March 6, 2013
Using DNA for data storage
March 5, 2013
You have all been doing that, of course -- storing your genome as DNA. That's not the point here, though it is part of the inspiration. The issue raised here is whether we might use DNA as something of a hard drive, or at least a tape drive, for storage of our computer data.
The incentive is simple... DNA is indeed a digital storage device. Each position in the DNA chain can be any of four bases, thus each position provides two bits of information. Knowing the mass of a DNA base, we can calculate that a gram of DNA could hold about 2x1020 bytes. (8 bits to a byte.) That's about 200 exabytes (EB), or 0.2 zettabytes (ZB). That's a lot of information -- and that's for just one gram of DNA. For perspective, it is estimated that the total amount of digital information in the world is about 3 ZB. 15 grams of DNA (about half an ounce) could hold all of the world's data. In principle.
It this worth considering? What's the catch? To address that, we need to be more specific, and address the strengths and weaknesses of DNA as a storage device, as well as the alternatives. A new article, from the European Bioinformatics Institute and the DNA technology company Agilent, attempts to address the issue. (Agilent is a northern California company that is a spin-off from Hewlett-Packard.) As part of their effort, they encode some computer files into DNA, and show that they can read the information. This establishes the basic technical feasibility of the plan.
So what are the strengths and weaknesses of DNA? One of the strengths was illustrated above: it is very compact. Another is that DNA is quite stable. We have noted work on retrieving genome information on DNA that is tens of thousand of years old. Now, not all old DNA is good. It depends on the storage conditions. If we make DNA for long-term storage, it will be stored under ideal conditions -- which is not difficult to do. Just dry it, and store it in a tightly capped vial away from light or heat or oxygen. Under such conditions, DNA will likely retain its information for thousands of years. Magnetic tape, a common current form of long term storage, lasts a decade or so.
On the downside, both reading and writing DNA storage are slow and expensive. There is no point in thinking about using DNA for the fast operations associated with a hard drive. As to cost, the cost of reading (sequencing) DNA has plummeted in recent years, as we have noted [link at the end]. The cost of writing (synthesizing) DNA has dropped, too. It's still a problem, but there is reason for optimism that further price reductions will occur.
With those strengths and weaknesses, it is reasonable to consider the possibility that DNA might be good for long term storage -- archival storage. Compact and stable. The relatively high cost of use gets amortized over a long time period. The analysis in the paper is largely directed at this use. The basic argument is that the cost of making (writing) the DNA is the dominant cost issue for DNA storage, and the cost of transferring tape every few years is the dominant cost for traditional tape storage. They make some simple assumptions about these, and calculate the trade-off. The following figure summarizes their findings. In principle, it's a simple trade-off, but they have crammed a lot of specifics onto this one graph.
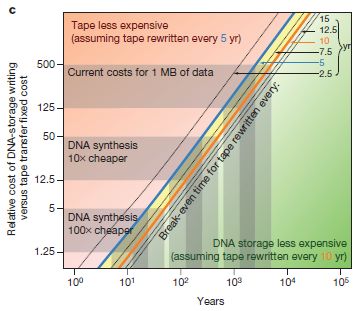
|
In this graph, the y-axis shows the cost of writing DNA. (It's actually the cost of writing DNA relative to the cost of transferring tape, but it is simpler to just think of it as the cost of writing DNA.) The x-axis is time. Note that both axes have log scales. The x-axis time scale goes from 1 year to 100,000 years. Each diagonal line is a trade-off line, where the costs of DNA and tape storage are equal. The various diagonal lines are for different intervals of refreshing the tapes; for a simple view, that turns out to matter little, so we can just look at those diagonal lines as a single block.
Here is the general picture... If the DNA cost is high, tape will be cheaper, except at very long times. If the cost of making DNA is reduced, then DNA will become cost effective at shorter times. The upper left part of the graph (reds) is a region of advantage-tape; the lower right (greens) is a region of advantage-DNA.
|
Let's look at an example... Consider 500 on the DNA-cost scale; that is about the current cost. Read across to the orange diagonal line: that is for refreshing tape every 10 years. Read down, and you will see that time is nearly 104 years. That is, with those numbers, DNA would be cheaper if you are considering long term storage -- on the order of 10,000 years. Now, go down to DNA cost = 5. Again, go over to the orange line (10-year tape refresh), and look down to the y-axis for the time: under 100 years. We're still talking about archives, but at least over time spans humans can comprehend.
In thinking about that previous point, remember that the specific numbers chosen were arbitrary. They chose to look at DNA cost = 5 because it is a simple 100-fold improvement, and they think it is a reasonable expectation. It is a guess how the cost of sequencing will decline; perhaps it will exceed their guess. Or perhaps it won't.
This is Figure 2c from the article.
|
What do we have here? A serious discussion of the possible use of DNA as a storage device for digital information. The numbers suggest that it might already be of some value for very long term storage. More importantly, the analysis suggests that with plausible cost reductions, it may become practical for the type of archival storage that is now common. It's an interesting challenge.
News stories:
* Researchers make DNA storage a reality. (Phys.org, January 23, 2013.)
* Store more. Much more! Big data meets tiny storage! (Why Files, January 23, 2013. Now archived.)
The article: Towards practical, high-capacity, low-maintenance information storage in synthesized DNA. (N Goldman et al, Nature 494:77, February 7, 2013.) It's not easy reading. I hope you will just find it interesting that serious work on the possibility of using DNA for storage of computer data is going on.
Those interested in how they implement the coding scheme should see the supplement, which is freely available at the article web site.
Intimidated by those less common metric prefixes? Check out my page Metric Prefixes - from yotta to yocto. The page includes examples to give a sense of scale for most of the prefixes.
An example of a post on ancient DNA: The Siberian finger: a new human species? -- A follow-up in the story of Denisovan man (January 14, 2011).
A post on the cost of DNA sequencing: The $1000 genome: Are we there yet? (March 14, 2011).
Also note: How long does DNA survive? (October 23, 2012). But caution, that post is really of little relevance to the current post, because survival depends critically on storage conditions.
There is more about sequencing on my page Biotechnology in the News (BITN) - DNA and the genome.
A post exploring another approach to long term data storage: Long-term data storage in glass(August 14, 2013).
More about data storage: Progress toward an ultra-high density hard drive (November 9, 2016).
Better violins through better fungi?
March 4, 2013
This is one of those stories that come up from time to time... a fascinating idea, with little information available. So we just briefly note it.
A Swiss group claims that treatment of wood with certain wood-rot fungi leads to violins that sound better. The fungi reduce the density of the wood. The idea is that reduction of the wood density leads to improved acoustical properties. If the fungal treatment is carefully controlled, this is done without undermining the structural integrity of the wood. Presumably much of the work involves figuring out the details. The news story refers to a test in which a violin made from fungi-treated wood was rated by listeners as better than a Stradivarius violin. See the video below.
The current news story is based on a talk by the lead scientist. We don't have much more to go on at this point. It's an interesting story, with interesting science behind it.
News story: Treatment With Fungi Makes a Modern Violin Sound Like a Stradivarius. (Science Daily, September 8, 2012.)
Video: Stradivari's Heirs - How Scientists Uncover the Secrets of the Stradivari. (YouTube, 4 minutes.) This is apparently a "trailer" for a film made in 2011 about the story.
The lab: Bio-engineered wood. From EMPA, in St Gallen, Switzerland. Scroll down to "Tonewood" and "Mycowood" for this project. Or choose the higher level heading "Applied wood materials" from the menu at the left for a broader view of the institute. It is an interesting site to explore.
More on violins:
* Briefly noted... The salt content of great 18th century violins from Cremona (March 16, 2022).
* Spiders and violins (May 4, 2012).
There is more about music on my page Internet resources: Miscellaneous in the section Art & Music. That section includes a list of related Musings posts.
More on fungi: SquarePants on Borneo (September 24, 2011).
More about wood density: Making wood stronger (March 19, 2018).
Also see... Using old clothes as building materials? (February 5, 2019).
More about rot: How fish rot -- and the implications for studying fossils (April 6, 2010).
CFL and LED lights: energy-efficient, but toxic
March 3, 2013
Incandescent bulbs (with a glowing filament) are out, and compact fluorescent lights (CFL) and light-emitting diodes (LED) are in. The major driving force for the change is energy. The newer light bulbs are much more energy-efficient than the old ones; incandescent bulbs turn much of their energy input into heat. However, the new bulbs have their own problems. We recently noted that CFLs may leak UV irradiation [see link at the end]. Further, the new bulbs use more metals.
A new report compares the use of metals by the various types of lights. There are two concerns here. One is that some of the metals may be toxic. The other is that they may be limited resources, and thus have supply and cost issues.
The following figure summarizes one of their analyses. In this case, the concern is toxicity to humans resulting from emissions to urban air.
|
The bar height shows the measure of hazard: the human-toxicity potential. Note that the scale is not only a log scale, but that the numbered marks are 1000-fold apart. Results are shown for various individual metals, and then for the total.
For simplicity, let's start with the "total" bars, at the right. There are three bars, one for each of the three types of lights, according to the color key at the bottom. The shortest bar is the blue bar, for incandescent bulbs. The bars for the other two types of bulbs are higher. It may not look like much, but they are about 100-fold higher.
|
If you look at the results for the individual metals, you will find that the general conclusion -- incandescent is lowest -- is true for almost every metal shown.
There is another observation that is interesting; you might be surprised by the result. Which metal is of the most concern? Browse the results for the various metals, and you will see it is zinc (Zn). That is true for each of the types of lights. Why is Zn of more concern than, say, lead (Pb)? Surely, Pb is more toxic than Zn? Indeed -- if you have the same amount of each. But there is much more Zn in the lights than Pb, so that the Zn effect is larger.
This is Figure 4a from the article.
|
The article contains several such analyses, in areas of human toxicity, ecotoxicity, and resource consumption. The broad pattern -- that CFL and LED lights are worse than incandescent lights -- holds for all the analyses. (The more specific point above, that Zn is the greatest toxic hazard, does not hold for all the analyses. Copper leads by some criteria, and aluminum and lead are also of concern.)
What are we to make of this? It's good information, and it should lead to improved products. The most important result of this type of analysis should be to spur the development of improved lighting products, with the energy efficiency of the new types but reduced hazards and resource consumption. Second, it is a reminder that the new lights should be disposed of properly. Accidental exposure to a broken CFL is not going to hurt you, but collectively, large numbers of these bulbs do represent a concern worth addressing.
News stories:
* Casting a shadow over green light bulbs. (Chemistry World, January 17, 2013.)
* New research shows CFLs and LED light bulbs have higher toxicity and resource depletion than incandescent bulbs. (Electronics TakeBack Coalition, January 16, 2013. Now archived.) The source here is an environmental organization; the quality of the information seems good, but always be cautious about accepting any single source.
The article: Potential Environmental Impacts from the Metals in Incandescent, Compact Fluorescent Lamp (CFL), and Light-Emitting Diode (LED) Bulbs. (S-R Lim et al, Environmental Science & Technology 47:1040, January 15, 2013.)
Background post: A possible hazard of using compact fluorescent light bulbs (November 13, 2012). This post deals with possible leakage of UV light from CFLs. Once again, this is a reminder that the new technologies are imperfect. They have benefits, but they are not fully developed.
Light bulbs (July 1, 2009). An earlier overview of lighting types.
Why might it be good to put lights on fish nets? (September 9, 2013). An application of LEDs.
More about lighting: Effect of artificial lighting on the environment (September 3, 2015).
My page of Introductory Chemistry Internet resources includes a section on Lighting: halogen lamps, etc.
Another life cycle analysis: Effect of food crops on the environment (November 20, 2015).
More about toxic things:
* Using wood-based material for making biodegradable computers (July 21, 2015).
* Is lipstick toxic? (July 2, 2013).
More about lead: Lead-rich stars (August 30, 2013).
Prospecting for gold -- with help from the little ones
March 1, 2013

|
At the left is one of the little helpers.
It is an Australian termite, Tumulitermes tumuli. It is about 6 millimeters long.
This is from the figure in the Phys.org news story. It was taken by the lead author of the paper.
|
In large parts of Australia, gold is found in commercially useful amounts a few meters below the soil surface. But finding it takes work. It requires drilling samples, and testing them, to see where the Au is.
It turns out that the termites that are so abundant there have already done the drilling step -- and deposited samples of the deep soil in their mounds. Why not, then, just measure the gold in the termite mounds?
|
This graph gives the idea. It shows the gold concentration (y-axis) for numerous locations (x-axis), as judged by two sampling methods.
The x-axis is a map position. There are no numbers on this frame, but there are tic marks; they represent 400 meter intervals. Thus the entire x-axis covers 2400 m. The samples studied were all on a line over this 2400 meter span. And that is what matters here: there is a line of samples.
|
There are two sets of data. The open symbols are for soil samples, from near the surface. The closed symbols are for samples from termite mounds. (The paper says that the termite mounds are on average about 50 m apart.)
The most prominent result is a peak in the curve based on samples from the termite mounds. The region of this peak is marked "mineralization" -- because it is known to be a region of higher Au levels. That is, high Au in the termite mounds marks regions of high Au n the deeper soil.
In contrast, the results for soil near the surface show little of interest. These samples may be easy to get, but they are not deep enough to be useful.
This is part of Figure 8 (the upper left frame) from the article.
|
This is an example of bio-prospecting: using some kind of biological sample as an indicator of minerals nearby. What's new here is the specific example. Although the work here is with termites that build substantial mounds, they suspect that many types of termites or ants would work. (If you haven't seen a termite mound, check out the Phys.org news story; the mounds are quite amazing.)
News stories. Some of the news stories on this work also refer to an earlier paper, in PLoS ONE in 2011, by the same group. That's fine, but I am focusing here on the new paper on gold.
* Termites and ants stockpile gold in their mounds, researchers find. (Phys.org, December 10, 2012.) Includes a picture of a termite mound (as well as the termite picture shown above).
* Termites Strike Gold: Ant and Termite Colonies Unearth Gold in Australia. (Science Daily, December 9, 2012.) The termites shown in this news story are a different kind of Australian termite.
The article: Source of anomalous gold concentrations in termite nests, Moolart Well, Western Australia: implications for exploration. (A D Stewart et al, Geochemistry: Exploration, Environment, Analysis, 12:327, November 2012.)
More about termites (and ants)...
* What if two of the world's most destructive pests spent the evening together in Fort Lauderdale? (April 4, 2015).
* Ants: nurses, foragers, and cleaners (May 24, 2013).
* Tracking termites (February 26, 2010).
More about gold...
* Added October 8, 2025.
Measuring the temperature of ultra-hot atoms -- and uncovering a new way to not melt gold (October 8, 2025).
* Goldene: 2-dimensional gold (July 9, 2024).
* Water disinfection in nanoseconds (April 3, 2023).
* A gold-based spray to treat heart damage (May 17, 2022).
* Air pollution: progress towards a process for ammonia oxidation (April 5, 2019).
* A simpler assay for detecting low levels of HIV, using gold nanoparticles (January 3, 2013).
* The golden ear: A nano-ear based on optical tweezers (July 13, 2012).
* Weighing gold atoms (September 24, 2008).
February 27, 2013
Can rats touch infrared light?
February 25, 2013
Of course not. Rats can't even sense infrared (IR) light.
But what if we give the rats an IR detector, and then wire it up to the part of their brain that detects touch? A team of scientists has now done just that, and the rats respond just as if they had touched the IR light.
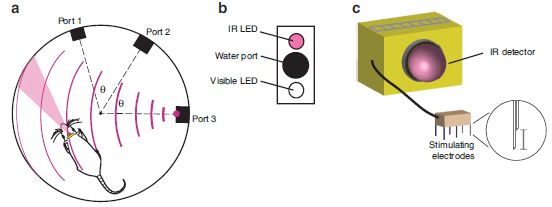
|
The figure above illustrates some parts of the set-up.
Part a diagrams the test chamber. The rat has an IR detector on its head. The triangular (or conical) pink region in front of the head shows where the sensor can see.
The test chamber includes three "ports". Part b diagrams one of the ports. It contains a water port, which provides a reward for the rat. It also contains two light-emitting diodes (LED), one emitting visible light and one emitting IR light. Going back to part a, you can see that port 3 is emitting IR.
Part c diagrams the IR detector that the rat is wearing. You can see that it is designed to plug in to something. In fact, it is plugged in to the rat's brain.
This is Figure 1 parts a-c from the article.
You can see the test chamber in the movies listed below.
|
The basic plan is that the rats are trained to associate the IR light with a reward, available at the water port. That this works shows that the rats receive the IR signal in the brain in a functionally useful form. But remember, the IR detector itself is an electronic device; what the rat gets is a specific brain stimulation as a result of what the electronic detector sees. The net effect is that the rat gains a sense -- a functional sense -- that is new to them.
The new sense was wired into the touch area of the brain; touch is a major sense for rats. They show that the rats still respond normally to touches of their whiskers. Thus the new sense seems to be integrated into the brain alongside an old sense, without disrupting it. I'm sure this will be the topic of much further work.
This is part of the broad area of studying the possible interfacing of electronic devices (and computers) with the brain. A novel feature here is that the rat gains a new sense, rather than simply having a defective one restored.
News stories:
* Brain implant gives rats a feel for infrared -- A sensory substitution device enables rats to perceive infrared light with their sense of touch. (Guardian, February 15, 2013.)
* Neuroprosthesis Gives Rats the Ability to 'Touch' Infrared Light. (Neuroscience News, February 12, 2013.) The end of this news page contains useful links. It links to the lab web page, which includes the movies listed below, with their descriptions; it also links to a copy of the article.
Movies. There are four short movie files posted with the article at the journal site listed below. You should be able to access the movie files whether or not you have subscription access to the article itself there. (The movies, with their descriptions, are also available via the Neuroscience News story, above.) Choose "Supplementary information", and scroll down to near the bottom of the page, where the movies are listed. The descriptions -- and the movies themselves -- are a bit cryptic, but at least you will get the idea of the test situation. It's worth looking through the four movies. The test in movie 3 is more difficult, because the ports are closer together; it is hard for the IR sensor -- and, therefore, the rat -- to distinguish the adjacent ports. In movie 4, a "blank" or "no stimulation" trial means that the sensor did not deliver a signal to the rat's brain. This served as a control that the rat's response to the IR light was through the sensor (and not through their eyes or some heat sensor).
The article, which is freely available: Perceiving invisible light through a somatosensory cortical prosthesis. (E E Thomson et al, Nature Communications 4:1482, February 12, 2013.) From this site, choose "Supplementary information", to get to the movie files noted above.
More from the same lab... Can one rat know what another rat is thinking? (April 8, 2013).
A post about an animal with a natural sense for infrared light: How to find the blood (August 29, 2011).
More about infrared light... Windows: independent control of light and heat transmission (February 3, 2014).
Another added sense... Can blind rats learn to use a geomagnetic compass? (June 29, 2015).
Also see:
* What if you had eyes on your tail? (July 27, 2013).
* Brain-computer interface: Paralyzed patients control robotic arm by their thoughts (June 16, 2012).
And see the accompanying post, immediately below; it also includes more links to related Musings posts.
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
Dung beetles follow the Milky Way
February 24, 2013
Dung beetles are fascinating insects. Their characteristic behavior is rolling dung balls across the African savanna. A recent post showed one aspect of thermoregulation by the beetles: they climb up on the dung ball to cool their feet [link at end]. Dung beetles can also travel at night. They travel in near-straight-line paths, getting their dung ball away from the crowd. They do so even on moonless nights -- if it is clear.
A new article suggests that the beetles can navigate at night by orienting to the Milky Way. This is based first on observations in the field. The scientists then follow up by taking the dung beetles to a planetarium, and observing their behavior there under controlled conditions.

|
A dung beetle with a cardboard cap -- to prevent it from seeing the stars.
One experiment reported in the new article shows that dung beetles wearing such a cap are unable to travel in straight paths at night. As one control, beetles wearing a transparent cap navigated fine.
This is trimmed and reduced from a figure in the SciTech Daily news story. It is probably equivalent to Figure 1C from the article.
|
Under the controlled conditions of the planetarium, the scientists could show that that the band of the Milky Way was the key sky feature for the beetles (in the absence of the much brighter moon). The beetles navigated well when the planetarium sky showed only the Milky Way, and navigated poorly when the sky showed only a collection of bright stars.
This is the first example of insects using the stars for navigation. It is also the first well-documented example of any animal using the Milky Way for navigation. I also suspect that the dung beetles studied here are the first to be treated to a planetarium show.
News stories:
* Study Shows that Dung Beetles Use Stars for Orientation. (SciTech Daily, January 25, 2013.)
* Dung Beetles Follow the Stars. (K Harmon, Scientific American, January 24, 2013.)
The article: Dung Beetles Use the Milky Way for Orientation. (M Dacke et al, Current Biology 23:298, February 18, 2013.) (Put the article title into Google Scholar, and you may find a copy of the article.)
More about dung beetles:
* Libyan desert glass, King Tut, and the hazards of meteorite strikes (May 31, 2019).
* What if dung beetles wore boots? (December 14, 2012).
More about beetles:
* How to preserve dead mice so they stay fresh and edible (January 18, 2019).
* How to fly a beetle (April 27, 2015).
More on the Milky Way:
* Could you find debris from a supernova in your backyard? (April 27, 2016).
* We are all Laniakeans (October 21, 2014).
* Mayhem at the center of the Milky Way (August 23, 2011).
A living organism powered entirely by electricity?
February 22, 2013
|
Cells of Mariprofundus ferrooxydans bacteria.
These are scanning electron micrograph (SEM) images.
This is Figure 2A from the article.
|
A striking feature is the unusual way these bacteria divide. As you can see from the one at the bottom left, they divide lengthwise, rather than across.
The more important point for now is how these bacteria are getting their energy. You can't really tell, but in the above figure the bacteria are attached to a graphite electrode. In the work reported in a recent article, the bacteria are being fed with an electric current -- which seems to be their sole energy source.
These bacteria normally grow by "burning" iron. More specifically, they oxidize Fe2+ ions to Fe3+ ions. This works, though it is a difficult existence. For one thing, the product Fe3+ precipitates out, covering the growing cells with rust. This makes it difficult to study these bacteria in the lab.
The scientists also knew that these bacteria would grow attached to a piece of iron; this suggested that their biochemical machinery for oxidizing the iron was, in part, on the outside of the cell. If so, perhaps they could get the bacteria to attach to a piece of graphite and then just feed them the electrons they would get by oxidizing the iron. They tried it, and it seems to work. The bacteria grow, apparently rather normally, but do not get coated with rust.
The iron or the electricity are the energy source for the bacteria. They still need other kinds of food, as "material". For carbon, they use carbon dioxide, "fixing" it just as plants do.
News story:
Scientists trick iron-eating bacteria into breathing electrons instead. (Phys.org, January 29, 2013.)
The article, which is freely available: Cultivation of an Obligate Fe(II)-Oxidizing Lithoautotrophic Bacterium Using Electrodes. (Z M Summers et al, mBio 4:e00420-12, January 29, 2013.)
Another example of unusual electron metabolism in bacteria: On sharing electrons (May 3, 2011).
More about iron chemistry... 2 + 2 = 4: Chemists finally figure it out (October 9, 2015).
February 20, 2013
Spiders in the sky
February 20, 2013
Look at the video, then read the news story.
Video. (YouTube, 5 minutes.)
News story: Why Thousands of Spiders Are Crawling in the Skies Over Brazil. (Wired, February 11, 2013.) The video is also included with this news story.
A recent post about another adaptation to the urban environment: Of birds and butts (February 2, 2013).
More about spiders from Brazil... Tarantulas in the trees (November 11, 2012).
More about spiders: Bat meets spider (March 29, 2013).
Of disasters, asteroids and meteors
February 19, 2013
Last week I juxtaposed two short items [links at end]. One was a news feature that discussed rare but inevitable major disasters, such as super-volcanoes. The other noted the fly-by of an asteroid later in the week -- a close encounter, but one of no real consequence, since the orbit of the asteroid was well established. The juxtaposition was intentional. However, there was no intent to imply that any significant event would actually occur. No intent. And no premonition.
Friday morning (the day of the asteroid fly-by) a headline news story was about a meteor strike in Russia, with some damage and several hundred people injured. (The number of injuries has since risen to about a thousand, mostly minor. The damage is estimated as several million dollars.) Two BBC announcers ("presenters", in BBC lingo) were arguing about whether it was related to the asteroid fly-by. They contacted an astronomer, who assured them it was not. Just a coincidence. The next morning, I learned we had had a meteor strike in northern California. Another one. We had two significant meteor strikes last year; one has already been the subject of an article in Science, because of the unusual composition. (None of the California meteor strikes noted here caused any injury or significant damage, so far as I know.)
What's going on? It's just the Universe doing its thing. In a sense it is what that news feature on disasters was about. Events happen. Small ones happen more frequently, bigger ones less frequently. Occasionally, there are super-disasters.
The asteroid that passed by -- the one event that was predicted -- was big enough that it could have devastated a city, perhaps injuring or killing millions. We only discovered it last year; had it been on path to hit Earth, we would not, at this point, have had any way to change its course. (We could have evacuated the target area.) The meteor that targeted Russia exploded before it hit ground; the damage was due to the shock waves, and the injuries largely due to people being hit with debris. In the big scheme, it was a minor event. To the people of the affected area, it certainly was significant, though not devastating. The three meteors that hit northern California in the past year were of similar size.
Here are some news stories about the meteor events; most are from the popular media. Remember that immediate reports of current events may not be entirely accurate.
Last Friday...
* Fireball Lights Up Northern California Skies: Reports. (Space.com, February 16, 2013.)
* Meteor Streaks Across Russian Urals, Leaves Nearly 1000 Injured. (Huffington Post, February 15, 2013; now archived.)
Last year, in northern California...
* Massive Fireball Over California Coast - October 17th, 2012. (American Meteor Society (associated with Pennsylvania State University), October 2012.) The page links to reports of recent meteor events -- many of them.
* Minivan-sized Asteroid Exploded Over California. (Discovery News (later, Seeker), April 23, 2012. Now archived.)
Background posts:
* Reasons to hide tonight? (February 11, 2013).
* An asteroid alert (February 11, 2013).
More about one of those northern California events noted above: Formation of the Moon: the California connection (October 10, 2014).
More about meteors...
* Libyan desert glass, King Tut, and the hazards of meteorite strikes (May 31, 2019).
* The origin of reactive phosphorus on Earth? (July 5, 2013).
More about asteroids: What has six tails -- and is beyond Mars? (November 20, 2013).
More to worry about... A visit from a star? (March 8, 2017).
There is no intent to post more on this topic next week.
Chocolate: 1200 years old
February 18, 2013
An intriguing story... Scientists have analyzed samples from 8th century human societies in what is now the US state of Utah. Chemical analysis of bowls points to large amounts of chocolate. It's the earliest evidence for chocolate in North America. The problem is that chocolate (from the plant Theobroma cacao) is not native to that area. Thus the result suggests a migration of chocolate from the more tropical regions (such as Mexico). That is a novel -- and surprising -- point, and many are skeptical.
Right now, there are more questions than answers, as often the case when a new discovery is made. The facts -- the basic chemical analyses -- may be right, but we may or may not be interpreting them correctly. For example, is it possible that the analyses -- of the specific chemicals theobromine and caffeine -- do not really point to chocolate? The authors address this, and tentatively reject it based on current knowledge, while recognizing that they actually know little about the native plants of the study area. Anyway, this new finding seems worth briefly noting.
News story: Earliest Evidence of Chocolate in North America. (Science Now, January 22, 2013.) Good overview of the work, and the controversy surrounding its interpretation.
The article: Cacao consumption during the 8th century at Alkali Ridge, southeastern Utah. (D K Washburn et al, Journal of Archaeological Science 40:2007, April 2013.)
More about chocolate:
* A better way to make chocolate, inspired by brake fluid (August 23, 2016).
* Better chocolate? Use better yeast? (May 3, 2016).
* Rats will free prisoners, and share their chocolate with them (January 18, 2012).
More about caffeine:
* Caffeine boosts memory -- in bees (April 12, 2013).
* Your desire for caffeine: It may be in your genes (May 31, 2011).
Sulfur dioxide in the atmosphere of Venus
February 16, 2013
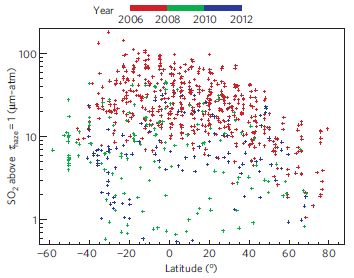
|
What an interesting graph! I chose to post this item partly because I was intrigued by this graph.
What does it mean? It shows the amount of sulfur dioxide, SO2, in the atmosphere of Venus as a function of latitude and time. SO2 is shown on the y-axis (log scale!) and latitude is shown on the x-axis (0° is the equator). Time is color-coded, according to the key at the top: for example, red points are the oldest measurements, from 2006 to 2008.
|
This is Figure 2 from the article listed below. The results are based on measurements taken by the Venus Express spacecraft, from the European Space Agency.
(The units for SO2 on the y-axis are complicated. What matters for now is that the numbers reflect the amount of SO2. "100" near the top means 100 times more SO2 than the "1" near the bottom.)
|
There is a clear pattern: red points are near the top. As noted, red points are the oldest points. Thus we see that the SO2 level on Venus has been decreasing since those earliest time points -- over a time scale of just a few years. You can see that simply from the color pattern of the points. (You can also see a trend toward higher SO2 levels nearer the equator.)
The results shown above, taken along with other data for SO2 in earlier years, suggests that the SO2 level on Venus fluctuates rather dramatically. Why? Well, they don't know, but one possibility is volcanic eruptions. Results such as those reported here may be the first evidence for active volcanism on Venus. Regardless of that interpretation, the work is impressive simply in the measurements it reports and the trends it finds.
News story: Have Venusian Volcanoes Been Caught in the Act? (Science Daily, December 3, 2012.)
Both of the following are freely available:
* News story accompanying the article: Planetary science: Rising sulphur on Venus. (L W Esposito, Nature Geoscience 6:20, January 2013.)
* The article: Variations of sulphur dioxide at the cloud top of Venus's dynamic atmosphere. (E Marcq et al, Nature Geoscience 6:25, January 2013.)
A post about atmospheric SO2 and volcanoes -- on Earth: Why isn't the temperature rising? (September 12, 2011). SO2 is a million time more abundant in the atmosphere on Venus than in ours.
More about volcanoes: Hawaii's hot spot(s) (October 9, 2011).
More about Venus: Venus: an unusual view (March 18, 2013).
More about atmospheric SO2 and volcanoes -- on Earth: SO2 reduces global warming; where does it come from? (April 9, 2013).
Planet Venus was also mentioned in these posts:
* GJ 1132b: "the most important planet ever found..." (December 18, 2015).
* A new trick for the Kepler planet-hunters (June 25, 2012).
* Collision of Earth and Mars (July 8, 2009).
Unusual synthesis of cadmium telluride quantum dots
February 15, 2013

|
Here is where they did it.
This is Figure 1a from the article.
The PAC is the posterior alimentary canal.
|
Cd2+ + Te2- --> CdTe (s)
It's simple chemistry. So simple an earthworm can do it.
CdTe (cadmium telluride) is a semi-conductor material. Small CdTe particles, of just the right size, known as quantum dots, are useful fluorescent particles. Engineers are finding uses for them in a range of applications, including LED lights. Biologists use them for labeling cell parts. However, they are not so easy to make. Chemists find it difficult to make CdTe of just the right properties. Earthworms seem to do it quite naturally.
This is one of those findings where the first reaction may be simply amusement. As the authors note, there certainly are precedents -- good reason to give it a try. So they try, and it works -- well enough that people may want to seriously consider its potential as a practical process.
A little more about what is going on... Apparently, earthworms are quite resistant to cadmium, which is generally considered a very toxic metal. In fact, what happens here is undoubtedly part of the worms' processes of detoxification. The worms concentrate the cadmium in a particular region of their digestive system. As to the tellurium... the authors feed the worms tellurite, TeO32-. The worms reduce it to telluride, Te2-. It's very much like an organism taking sulfate or sulfite ions (SO42- or SO32-) and reducing them to sulfide (S2-), which the organism uses. Bringing the cadmium and telluride ions together leads to the formation of the CdTe; little is understood about the details.
News stories:
* Researchers use earthworms to create quantum dots. (Phys.org, December 28, 2012.)
* The Quantum Earthworm. (Carl Zimmer, The Loom (National Geographic blog), December 24, 2012.)
Both of the following are freely available:
* News story accompanying the article: Nanomaterials: Earthworms lit with quantum dots. (R D Tilley & S Cheong, Nature Nanotechnology 8:6, January 2013.)
* The article: Biosynthesis of luminescent quantum dots in an earthworm. (S R Stürzenbaum et al, Nature Nanotechnology 8:57, January 2013.)
Another "biological" approach to making quantum dots: Using an enzyme to make quantum dots (January 6, 2023).
More about cadmium-based nanoparticles, quantum dots, and such:
* Using light energy to power the reduction of atmospheric nitrogen to ammonia (May 20, 2016).
* A more powerful method for measuring what is in a cell (July 23, 2013).
More cadmium: Data on 31 isotopes of cadmium support a new model for the structure of atomic nuclei (December 1, 2018).
More about toxic metals: Is lipstick toxic? (July 2, 2013).
Another Annelid worm... A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
Also see: Xystocheir bistipita is really a Motyxia: significance for understanding bioluminescence (May 9, 2015).
February 13, 2013
Cockroach should be disinfected before eating it
February 12, 2013
You probably spend time cleaning your food. But did you know... Parasitic wasps that eat cockroach clean their food, too. A new article reports that larval wasps, living inside the cockroach, secrete a potent antibacterial mix.
|
This shows a wasp larva inside a cockroach. The arrows point to droplets of larval secretions.
The secretions contain antibiotics (anti-bacterial agents). The wasp larva spreads the secretions around before eating.
This is Figure 1 from the article. It is a frame from the movie file noted below.
|
How can we see what is going on inside the cockroach? Because the scientists had opened it up, and inserted a clear window (a coverslip) into the roach body.
Based on their evidence, the scientists believe that the wasp secretes the antibiotics to clean its food. Their story is incomplete: they do not directly show that the secretions are necessary for the larval survival. It would be interesting if they can figure out a way to test that.
News story: Researchers discover wasp larva disinfect their food before eating. (Phys.org, January 8, 2013.) A good overview of the work, with a description of the larval life cycle. (It includes a picture of the adult wasp. It's a quite pretty little critter, perhaps as suggested by its common name, the emerald cockroach wasp.)
The article: Larvae of the parasitoid wasp Ampulex compressa sanitize their host, the American cockroach, with a blend of antimicrobials. (G Herzner et al, PNAS 110:1369, January 22, 2013.)
Movie. There is a movie posted with the article. It shows a wasp larva inside the cockroach, and shows the larval secretions. It's not dinner-time viewing, but it is interesting. Go slowly at the start, so you figure out what is what; it is well-labeled, but may move faster than you want. The movie runs about 30 seconds. From the article web site, above, choose Supporting information. The movie is also available at movie: Ampulex compressa sanitize their host. (YouTube)
More on parasitic wasps...
* The benefit of providing alcohol to the eggs (March 30, 2013).
* Wasp hides under ladybug (January 3, 2012).
More on cockroaches:
* Why and how some cockroaches avoid glucose (October 11, 2013).
* Acrobatic cockroaches inspire robot design (September 16, 2012).
The post started with a comment about keeping your food clean. If you need a reminder why... Killer chickens (December 2, 2009) (and several posts linked there).
More on antibiotics and disinfectants is on my page Biotechnology in the News (BITN) -- Other topics under Antibiotics.
Reasons to hide tonight?
February 11, 2013
A supervolcano might erupt -- and wipe out life on a continent scale, or worse. And there are some other concerns. A recent news feature in Nature discussed some of them. It's a couple pages, and much fun. After all, none of the super-disasters they present are likely to happen soon. Or are they?
News story, freely available: Planetary disasters: It could happen one night. Catastrophes from the past will strike again -- we just do not know when. (N Jones, Nature 493:154, January 10, 2013.)
More about volcanoes:
* Hawaii's hot spot(s) (October 9, 2011).
* VPOW (July 14, 2010).
And see the accompanying post, below.
There is also a post that is something of a follow-up to both of these:
Of disasters, asteroids and meteors (February 19, 2013).
The back-door gene?
February 9, 2013
Imagine that you are going to build yourself a house. Would you include a back door? Do you think that your choice about a back door is determined by a gene? That is, do you have a gene for a back door?
A new article shows that mice seem to have such a gene, as well as other genes that determine how they build their burrow -- or house.
Here is the idea. The scientists study two closely-related species of mice. One species forms burrows with an escape channel (or back door), one does not. In the lab, both species make burrows as expected. Importantly, they make their characteristic burrows even if they are "naive" -- have never seen another of their kind make a burrow. That is, the burrow type seems to be innate, not learned.
They then cross the two types of mice. All of the progeny, the F1 in traditional Mendelian terminology, make escape channels. This is the classic result expected when a single gene is involved, with one form being dominant. Further, they cross the F1 back to the non-escape-channel parent strain; half the progeny make escape channels. Again, this is what is expected if there is a single gene involved, with escape channels being dominant.
Those who have had some genetics can draw out these crosses; remember that the F1 mice are all heterozygous, and the backcross is to the homozygous recessive parent.
Thus the simple crosses suggest that a single gene is involved is determining that one species makes an escape channel and one does not. We should caution this does not mean only one gene is involved in the behavior; it means that the two species studied here differ in one gene. Further, it does not exclude some more complex explanations, such as there being multiple genes that are closely linked on the chromosome. The logical point of the work is that it points towards a single gene difference.
The two mouse species also differ in the length of their primary burrow channel. The genetic analysis for this is more complex, but suggests a fairly small number of key genes each contributing "a few centimeters" to the channel. Once again, the work points to a limited number of genes that seem to somehow control complex behavioral traits.
News stories:
* Mouse burrowing 'in their genes'. (BBC, January 17, 2013.)
* Complex behaviors driven by remarkably simple genetics -- A mouse's intricate architectural tastes are the product of "modular" genetics. (Ars Technica, January 16, 2013.)
* Behaviour genes unearthed -- Speedy sequencing underpins genetic analysis of burrowing in wild oldfield mice. (E Callaway, Nature News, January 16, 2013. Also in print: Nature 493:284, January 17, 2013.)
Movie. There is a movie posted with the article. The first scene shows a mouse escaping its burrow via the back door (escape channel) when a "snake" enters from the front door. Note that the back door is not visible before the mouse exits. The escape channel ends just below the surface -- close enough that that the mouse can easily escape. You may need to play this segment more than once to appreciate it; things happen fast. The second scene extends to the end, and may mystify you for a while. Be patient, and you'll get to see the burrow -- a foam cast of the burrow. The movie is freely available at the article web site (below) -- whether you have subscription access or not. It is also included with the Ars Technica news story (above).
* News story accompanying the article: Evolutionary genetics: Genes for home-building. (P Goymer, Nature 493:312, January 17, 2013.)
* The article: Discrete genetic modules are responsible for complex burrow evolution in Peromyscus mice. (J N Weber et al, Nature 493:402, January 17, 2013.)
Other posts about home building include...
* Of birds and butts (February 2, 2013).
* What if your house could sweat when it got hot? (November 30, 2012).
The role of the immune system in making stem cells
February 8, 2013
An intriguing and confusing story -- one that may turn out to be important.
Some background. One major type of stem cell being studied is the induced pluripotent stem cell (iPSC). iPSCs are made from various types of body (somatic) cells by reprogramming them to an embryonic-like stage, where they are pluripotent: capable of making any cell type. The basics for making iPSC were developed in 2006 by Shinya Yamanaka, who shared the Nobel prize last year for this work.
To induce the reprogramming of adult cells to become embryo-like requires adding some "factors". The original way to do this was to add a virus that coded for the needed factors. This works, but using the virus raises some concerns. Therefore, people developed other ways to deliver the factors, without a virus. Such virus-free procedures work, but not very well. Why a virus-free procedure works poorly has been mysterious. And that leads us to the new work.
The key experiment in the new work is to use a non-viral method of inducing reprogramming -- but also add a virus, a "blank" virus, with no relevant genes. Turns out, adding the virus improves the efficiency of reprogramming. That is, even though all known needed factors were provided independently of the virus, there was something about adding a virus that improved the procedure.
|
This figure gives an example of what they did.
The general plan here is to add Sox protein, and see how much Nanog protein is made. Sox is one of the factors used to induce iPSC; Nanog is a protein associated with making pluripotent stem cells. That is, higher level of Nanog is "good" in this context. The level of Nanog is shown on the y-axis, as relative amount.
The three curves differ in how Sox was supplied. That is the point of the experiment.
|
The cryptic labeling makes the details hard to decipher, so I need to explain what those various methods were. One way to approach this is by looking at the results. Two methods worked: a lot of Nanog was made. One method did not work: little Nanog.
Start with the red and blue curves. Both involved delivery of Sox -- by different methods. The red curve (top) used delivery of Sox by a virus; this worked. The blue curve (bottom) used delivery of Sox as a free protein; this did not work. Then we have the green curve... In this case, they delivered Sox as a free protein, but also added a virus. The virus had no relevant genes, but it was a virus. Adding this "blank" virus made the free protein work. That is, if you compare the blue and green curves... In both cases, Sox was delivered as a free protein; with the green curve, a blank virus was added, and that made it work.
This is Figure 1A from the article.
|
The authors go on to show that the virus seems to be acting by stimulating the immune system. More specifically, it is acting through one of the toll-like receptors (TLR) of the innate immune system. Thus, the original Yamanaka success seems to have involved not just the identified factors, but also the virus itself, via the innate immune system. This is a small step toward better understanding the reprogramming process. I'm sure we will be hearing more about this and other developments. It's a reminder that the iPSC process is still new, and only partly understood.
News story: Viruses Affect Cell Reprogramming -- Viral vectors used to carry transcription factors that de-differentiate cells into a stem-cell-like state are themselves a key factor in efficient reprogramming. (The Scientist, October 25, 2012. Now archived.)
* News story accompanying the article: ''Transflammation'': When Innate Immunity Meets Induced Pluripotency. (L A J O'Neill, Cell 151:471, October 26, 2012.)
* The article: Activation of Innate Immunity Is Required for Efficient Nuclear Reprogramming. (J Lee et al, Cell 151:547, October 26, 2012.)
For more on stem cells:
* Added April 23, 2025.
The choano-mouse: implications for understanding the origins of transcription factors that control stem cells (April 23, 2025).
* Bacteria can make mouse stem cells (April 6, 2013).
* Geron sells its stem cell business (January 23, 2013). The business side -- of products based on embryonic stem cells.
* Using patient-specific stem cells to study Alzheimer's disease (February 24, 2012). An example of how iPSC are used to study disease.
There is more on stem cells on my page Biotechnology in the News (BITN) - Cloning and stem cells.
More on the innate immune system and TLRs: Why mice don't get typhoid fever (November 26, 2012).
More about immune systems: Bach and the immune system (August 26, 2013).
February 6, 2013
An improved CT scanner, with a reduced radiation dose
February 5, 2013
Briefly noted...
The NIH news release listed below describes an interesting development: an improved scanner for computed tomography (CT). It exposes the patient to less radiation, while providing improved images. Such progress is technology, of course. I have not seen the paper behind this, but it seems worthwhile noting this advance. Over time, there will be further experience with the new scanner, which will further show its benefits and weaknesses.
News story: Next-generation CT scanner provides better images with minimal radiation. (NIH, January 31, 2013.) This provides a good overview of how the scanner is improved. It also summarizes the findings reported in the article. It is from the funding agency, the US National Institutes of Health.
The article: Submillisievert Median Radiation Dose for Coronary Angiography with a Second-Generation 320-Detector Row CT Scanner in 107 Consecutive Patients. (M Y Chen et al, Radiology 267:76, April 2013.) I do not have access to this, and have not seen it. There is an extended abstract available.
More about radiation...
* Measuring radiation: The banana standard (April 17, 2011).
* Does radiation treatment of cancer cause new cancers? (April 8, 2011).
My page of Introductory Chemistry Internet resources includes a section on Nucleosynthesis; astrochemistry; nuclear energy; radioactivity. That section contains some resources on the effects of radiation.
A new SARS-related virus seems to be emerging -- and an "ethics" story
February 4, 2013
We have noted various new viruses [links at end]. One point of such stories is to allow us to watch the course of a new virus. We even hope that our societies (and public health institutions) learn from each new virus story so that we are better able to deal with one that may be serious; we must emphasize that in the early stages we have little way to know which will be serious. We now have another new virus to watch -- and it is killing humans.
Some of you may remember the SARS virus, which emerged in 2002. Fortunately, the SARS virus was not easily transmitted from one person to another, and good containment procedures brought the SARS epidemic to an end. SARS was on the scene for less than a year, but killed over 700 people -- 10% of those known to be infected -- during its brief stint.
We now have a new virus, which first drew attention in September 2012. It has killed five people so far -- five confirmed deaths, out of nine known to be infected. The virus has been isolated, and is undergoing study; it is a corona virus, the same general type as the SARS virus. This post is prompted by one recent paper on the new virus. The point is not so much the specific findings, but to bring the story of the new virus to your attention. We will watch it over the coming months. Will this new SARS-type virus do more or less damage than the original SARS? If it does do only limited damage, is that because we did something good to stop it, or because it just wasn't a very good virus?
What is in the new paper? One thing they tested was whether the new virus uses the same receptor as the SARS virus. (The receptor is the surface structure on cells where the virus attaches.) Simple studies show that it does not; for example, cells without the SARS receptor can be infected by the new virus. That's disappointing; if it used the same receptor, we would have a head start in knowing how it infects. Secondly, they tested the host range of the new virus: what kinds of cells can it grow in? Many. It grows in cells from a wide range of animals.
News stories:
* New SARS-Like Virus Infects Both Human and Animal Cells. (Science Now, December 11, 2012.)
* New coronavirus can infect cells from multiple species. (CIDRAP, December 11, 2012.) See links on the page to "MERS-CoV" or "SARS" for more, including recent news. MERS stands for the newly established name for the virus, Middle East Respiratory Syndrome.
The article, which is freely available: Human Coronavirus EMC Does Not Require the SARS-Coronavirus Receptor and Maintains Broad Replicative Capability in Mammalian Cell Lines. (M A Müller et al, mBio 3:e00515-12, December 11, 2012.)
The "ethics" story? Viruses can be the subject of international disputes. We have noted a previous example [link at end]. The new virus is raising its own international issues. It was isolated in one country, and then sent to a leading lab in another for further characterization (including the work discussed above). This has lead to concerns and questions. Among them is the tension between the scientist who isolated the virus and the authorities in the country where that happened.
As you read the following news story... The purpose here is not to blame anyone. The issues raised need to be addressed. The previous dispute was resolved by an international agreement, mediated by the World Health Organization (WHO). We hope further agreements will be worked out to deal with situations such as this new one. The goals should be clear: efficient development of understanding of new diseases -- which do not respect international boundaries. The concerns of developing countries are often understandable, and need to be respected -- without endangering public health anywhere.
News story: Tensions linger over discovery of coronavirus. (Nature News, January 14, 2013.)
Background posts about new viruses:
* The Heartland virus (October 2, 2012)
* Schmallenberg virus (January 20, 2012). Links to follow-up.
Follow-up for the new MERS virus: Where is the MERS virus coming from? (September 22, 2013).
More MERS... MERS in the United States (May 18, 2014).
The previous international microbiology dispute, and its resolution: International relations: sharing flu viruses (May 28, 2011).
Influenza is a continuing story of new viruses. Many posts on various flu issues are listed on the supplementary page: Musings: Influenza.
Two sections of my page Biotechnology in the News (BITN) -- Other topics are relevant to the virus story here. One is specifically on SARS, MERS (coronaviruses), and one is on the more general topic of Emerging diseases (general).
There is a section of my page Biotechnology in the News (BITN) -- Other topics on Ethical and social issues.
Of birds and butts
February 2, 2013
Urban birds may use cigarette butts as nest-building material. Nests with butts of smoked cigarettes have reduced numbers of parasites. Here is the data...
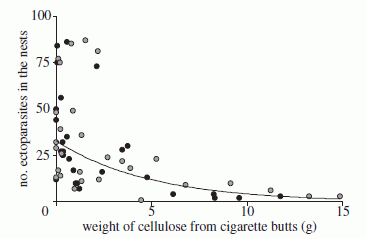
|
The graph shows the number of parasites (such as mites) found in nests vs the amount of cigarette butt material.
There is clearly a trend: more butt, fewer parasites.
This is Figure 1 from the article listed below.
|
What does this mean? They have no direct observations of the nest building process, so do not know how the birds choose nest materials. However, it is known that birds such as these do tend to choose plant materials with odors -- odors that probably correlate with insecticidal properties. Cigarette butts contain nicotine, which is certainly an insecticide. Thus it is plausible that including cigarette butts is a good choice by the birds, protecting the nest against parasites.
They have no information on whether there may be any harmful effects of the butts on the birds. That would apply also to any choice of nest materials.
The first reaction to an article such as this may be amusement. But there are serious biological questions here. Among them... How do birds choose nest materials? Is health one factor? If so, how do they do it? And how do birds adapt to new environments -- including man's urban environment?
News stories:
* Marlboro Chicks -- Two species of songbirds pack their nests with scavenged cigarette butts that repel irksome parasites. (The Scientist, December 5, 2012.)
* Cigarette Butts in Nests Deter Bird Parasites. (Scientific American, December 4, 2012.)
The article: Incorporation of cigarette butts into nests reduces nest ectoparasite load in urban birds: new ingredients for an old recipe? (M Suárez-Rodríguez et al, Biology Letters 9:20120931, February 23, 2013.)
Posts about birds include...
* Are urban dwellers smarter than rural dwellers? (August 2, 2016).
* A bird nest (September 9, 2014).
* Airport food: What do the birds eat? (May 24, 2014).
* The effect of cars on birds (August 2, 2013).
* How long does DNA survive? (October 23, 2012).
* The story of the peppered moth (July 9, 2012).
* Why don't woodpeckers get headaches? Designing better shock absorbers (April 18, 2011).
* Bird lays egg (March 19, 2011).
* Bird theater (October 19, 2010).
* Complex tool use by birds (May 28, 2010).
* Dancing birds (May 6, 2009)
Other posts about home building include...
* How did a one-ton dinosaur incubate its eggs? (July 13, 2018).
* The back-door gene? (February 9, 2013).
A post about another adaptation to the urban environment: Spiders in the sky (February 20, 2013).
More about butts: Butt batteries (December 16, 2014).
A better understanding of the basis of color vision
February 1, 2013
|
The color of some artificial visual pigments made by scientists at Michigan State University.
This is part of Figure 3 from the article listed below.
|
We'll come back to the figure in a moment, but first let's look at some background. The basis of our visual system is the absorption of light by a photoreceptor pigment in the eye. The pigment has two parts. One is retinal, a derivative of vitamin A; the other is a type of protein called opsin. Retinal binds to the opsin; this bound complex is the photoreceptor.
It is the retinal part of the photoreceptor complex that absorbs the light. But when you look at the various kinds of photoreceptors, such as the three color receptors in humans, they all have the same retinal; it is the opsin protein that varies. Somehow the chemical environment in the various opsin proteins changes the absorption spectrum of the bound retinal. Exactly how this happens has not been clear.
That gets us to the new work. The scientists set out to explore that effect of chemical environment on the color. It turns out that opsins are fairly difficult proteins to work with in the lab, so they used something else: another retinal-binding protein. They made a range of changes in the protein, by introducing mutations into the gene. They then measured the absorption spectrum of the complex of each of them with retinal. The paper contains full spectrum information, but the photos above give the idea -- very well.
In the figure above, M1 through M11 are the various opsin proteins they made. In each case what you are seeing is the color of a solution containing the indicated protein with bound retinal. There is quite a range of colors. The first tube contains retinal, with no protein at all. It appears colorless to our eye; actually it does absorb light, but only in the ultraviolet part of the spectrum. The full figure in the article contains the full spectra, and a table showing the absorption maximum for each species, as well as for natural visual pigments.
For example... Protein M1 absorbs blue light, with its maximum absorbance at 425 nm. This is very close to the blue photoreceptor for humans. By absorbing blue light, the solution of the protein appears yellow.
The red receptor M11 has an absorbance maximum at 644 nm. That's far beyond our red receptor, and beyond any retinal-based photoreceptor seen in biology. In fact, it is more red-shifted than many thought would be possible.
By making all these proteins and seeing how each one influences the absorption spectrum of the retinal, they learn more about how receptors for color vision work.
News story: Detailing Color Vision -- Scientists engineer a spectrum of artificial pigments to understand how animals see in color. (The Scientist, December 6, 2012. Now archived.)
* News story accompanying the article: Biochemistry: Redder Than Red. (T P Sakmar, Science 338:1299, December 7, 2012.)
* The article: Tuning the Electronic Absorption of Protein-Embedded All-trans-Retinal. (W Wang et al, Science 338:1340, December 7, 2012.)
The role of opsin proteins and retinal (vitamin A) in vision was noted in the post An unusual eye? (June 6, 2012).
And more...
* The eyes of Cnidaria (jellyfish): the big picture (August 19, 2018).
* Color vision: an overview (December 1, 2014).
* How can the mantis shrimp see so many colors of UV? They use filters (August 30, 2014).
A source of dietary vitamin A was discussed in the post Golden rice as a source of vitamin A: a clinical trial and a controversy (November 2, 2012).
Vitamin A (and hence retinal) is made from carotenoids. Carotenoids were featured in the post Red and green aphids (June 2, 2010). That post includes some information on the structures of various carotenoids and how they are made. Cut a molecule of beta-carotene in half, and you get, with just a bit of adjustment, two molecules of vitamin A.
More about vision: What if there was a gorilla in the X-rays of your lungs? (July 26, 2013).
Also see a section of my page Internet resources: Biology - Miscellaneous on Medicine: color vision and color blindness.
January 30, 2013
Monogamy
January 30, 2013
This story starts with a picture that caught my attention:

|
What do you think this is?
This is trimmed from the figure in the Live Science news story listed below. It's trimmed to focus on the face.
|
It's an owl monkey, a small monkey mainly from South America. It's nocturnal -- as you might guess from the eyes; that is an uncommon trait for monkeys.
The owl monkey is also monogamous -- another uncommon trait for monkeys. In a new article, scientists report studies of a natural population of owl monkeys. A key finding is that those that remain monogamous have more offspring than those that change partners -- an event that occurs most often due to a violent intrusion.
It's an interesting story. Simply studying such a population in nature is an achievement. Their discussion of some of the underlying issues is interesting, even if speculative at times. The quality of the data is not all that great -- no big surprise for something as complex as this. In any case, resist the temptation to generalize too much on the findings. But do enjoy the picture -- and read over one of the news stories for the ideas.
News stories:
* Monogamous Owl Monkeys Have More Babies. (Live Science, January 23, 2013.) This is the source for the figure shown above, and it gives a useful brief overview of the work.
* Owl Monkeys Who 'Stay True' Reproduce More Than Those With Multiple Partners. (Science Daily, January 23, 2013.) This is a more complete description of the work. At the end, it links to the article.
The article, which is freely available: Till Death (Or an Intruder) Do Us Part: Intrasexual-Competition in a Monogamous Primate. (E Fernandez-Duque & M Huck, PLoS ONE 8(1):e53724, January 23, 2013.)
More about monogamy: Is the hormone of love also the hormone of discrimination? (January 29, 2011).
Another primate with big eyes... Tarsier; eukaryotic cells (August 31, 2009)
If terms like owl monkey intrigue you -- or confuse you -- why not try Quiz: The monkey-cat (October 26, 2011).
More on monkeys: Pink corn or blue? How do the monkeys decide? (June 9, 2013).
Also see: When did mammals take on daylight? (February 11, 2018).
POCDx -- What's the barrier?
January 29, 2013
The first barrier for some might be, what does POCDx mean? It means point-of-care diagnostics; the simpler form POC is also used. POC is about providing medical testing that can be done on-the-spot, with fast and inexpensive results. Sort of.
Much work is being done to develop POCDx; there is a POCDx group at Berkeley. But getting POCDx to work requires more than making clever inexpensive tests; they must be integrated into the medical system. A talk at a Berkeley symposium in early January emphasized that POC was not just about technology but about the broader problem of health care systems. The talk was by Madhukar Pai, an India-trained doctor now at McGill University, and it was one of the high points of the day-long meeting. Pai noted that he had recently written an article on the topic -- and that is the point here.
The article, which is freely available: Point-of-Care Testing for Infectious Diseases: Diversity, Complexity, and Barriers in Low- And Middle-Income Countries. (N P Pai et al, PLoS Medicine 9(9):e1001306, September 4, 2012.) (Since I mentioned that our speaker was M Pai... The lead author, whose name is listed here, is his wife, who is also a physician at McGill.) Worth at least a browse for anyone interested in health care systems. And it is relevant to all, not just developing countries. The cost of health care in the US is a serious problem; POCDx offers one line of hope for improvement in our inefficient system. (Some have joked that India will lead in the improvement of US heath care, for this reason. We don't need to take that too literally, but it expresses a useful idea.)
More on health care:
* High-tech medicine: good or bad? (June 4, 2010).
* Maternal mortality (May 7, 2010).
* Healthcare costs (June 18, 2009).
Examples of POC developments:
* Ultra-fast PCR (March 14, 2023).
* The paperfuge: a centrifuge that costs 20 cents (April 17, 2017).
Fructose and your brain
January 28, 2013
Musings has discussed the fructose issue from time to time [link at the end]. A key question is what role fructose plays in the development of obesity.
A new article makes an interesting contribution to the fructose story. The basic plan... The scientists fed their test subjects a drink containing either glucose or fructose, and then did functional magnetic resonance imaging (fMRI) scans of the brain. The same subjects were tested with both sugars.
|
Here are some of their results...
The y-axis of the graph shows the blood flow change found by the fMRI test. "CBF", shown on the axis label, means cerebral blood flow. This is the basic parameter measured by fMRI. The x-axis is time after consuming the sugar. Results are shown for consuming fructose (triangles) and glucose (circles).
It is clear that consuming glucose and consuming fructose lead to different results.
This is Figure 1A from the article.
|
More specifically, the result shown here is for the hypothalamus area of the brain, which is involved in controlling appetite. The glucose leads to reduced appetite -- and to reduced activity in the brain region that controls appetite.
Caution... The previous paragraph may have suggested something, but there are various reasons why whatever it suggests should be taken with great caution at this point. Both the article and the accompanying editorial emphasize that this should be considered a preliminary result. The test situation is artificial: consuming a drink containing a large amount of the sugar. Further, the brain scan is not very high resolution. The authors consider this work as something like a proof of principle. Under well-defined special conditions, they can see this difference between the two sugars. It now remains to be seen what this means under relevant conditions. That is, this work does not answer any particular question about fructose, but it may provide a new tool to help us understand it.
News story: Fructose Has Different Effect Than Glucose On Brain Regions That Regulate Appetite. (Science Daily, January 1, 2013.) At the end, this item links to the article; in turn, the article and editorial link to each other.
* Editorial accompanying the article: Fructose Ingestion and Cerebral, Metabolic, and Satiety Responses. (J Q Purnell & D A Fair, JAMA (Journal of the American Medical Association) 309:85, January 2, 2013.)
* The article: Effects of Fructose vs Glucose on Regional Cerebral Blood Flow in Brain Regions Involved With Appetite and Reward Pathways. (K A Page et al, JAMA 309:63, January 2, 2013.)
Background post... Fructose; soft drinks vs fruit juices (November 7, 2010).
More about appetite: YY in the mouth? (April 4, 2014).
Another post about fMRI: Dog fMRI (June 8, 2012).
My page Organic/Biochemistry Internet resources has a section on Carbohydrates. It includes a list of related Musings posts.
Fish with bigger brains may be smarter, but ...
January 25, 2013
As background, humans are noted for having a large brain, but that brain is expensive. The human brain is about 2% of the body, but uses about 20% of the energy. Interestingly, the total energy consumption by humans is about what one would expect for the body size. Something must be compensating for the increased energy consumption by the brain. This issue has long been recognized, but is not entirely understood. One suggestion is that humans have less digestive system, compensating for the greater brain. Some have even suggested that the key to developing a bigger brain was learning to cook food, making it easier to digest. Some of this is little more than speculation at this point.
Now the fish. A team of scientists has decided to look at the big-brain issue with guppies. It's an interesting story; however, it is not at all clear what we should conclude from it at this point.
Here is an overview of what they did... They selected for fish with larger brains. (In parallel, they also selected for small-brained fish.) The resulting fish not only had larger brains, but were smarter -- as judged by how they did on a math test. That is, the brain was not only larger but better. The downside? The large-brained fish had smaller guts and produced fewer offspring.
Let's look at some of the specifics...
The work started by selecting for large-brained fish. This is logically straightforward, and turned out to be easy. They took the fish with the largest brains, and bred them. In parallel, they did the same for small-brained fish. Interestingly, over just two generations, they developed two populations of fish with 5-10% difference in relative brain size. That they did this so easily means they were largely exploiting natural genetic variation within the population.
Here is what they found in one test comparing the two populations...
|
This is a test of cognitive abilities of the small- and large-brained fish (red and blue bars, respectively). Results for females (left) and males (right) are shown separately.
For the females (left pair of bars), the large-brained fish (blue bar) do substantially better. For the males, no difference is seen. This is an interesting point, possibly even important -- and it is not mentioned further.
It's also of note that random guessing by the fish would give a score of 4 on this test. Three of the bars on the graph are approximately 4. The only bar different from 4 is the one for large-brained females. I must say I'm uncomfortable with this. The results suggest that a new skill emerged, rather than was enhanced. I'd really like to see more data on this point.
This is Figure 2 from the article.
|
And now some data showing the cost of having the larger brain...
The graph at the right shows the number of offspring from the small- and large-brained fish. Small-brained fish have more offspring. (The size of the offspring was the same.)
This is Figure 3B from the article.
In other work in the article, they show that the large-brained fish have smaller digestive systems.
|
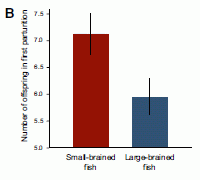
|
What do we learn from this? The first point is simply that they can do all this -- that they can select for larger (and smaller) brains, and study the effects. That is, they seem to have developed an interesting experimental system. Do the results here tell us anything about humans? Do they even tell us much about fish?
What they did here seems logical, but there are many questions about the work. I've noted some along the way, but I can think of more... They do this one "math" test to show that the fish with larger brains are smarter. Are their results here really significant, or was this some kind of fluke result? What other abilities would these fish show -- or not show? They test their small- vs large- brained fish; what about the original "normal" fish? (Maybe the large-brained fish test the same as normal, and the small-brained fish are odd.) Even if the fish chosen for large and small brains by this procedure have certain features (intellectual and otherwise), is that general? What would happen under different conditions? What would happen if these fish were bred for extended periods with limiting food, where a smaller gut might be a problem?
I don't have answers to these questions. That's the point. What they did here was interesting; the results are intriguing. But let's be cautious about interpreting this. It is hard to know what the significance is -- without much more work. The good news is that the fish system is simple enough and fast enough that they should be able to do more work. Let's see what bigger story -- and bigger brain -- develops.
News story: Big Brains Are Pricey, Guppy Study Shows. (Science Daily, January 3, 2013.) The story is a good description of the work, but I think it is overly generous in stating the significance. The logic is fine. However, I really see this as more of an initial effort along a new and interesting line than anything conclusive at this point.
The article: Artificial Selection on Relative Brain Size in the Guppy Reveals Costs and Benefits of Evolving a Larger Brain. (A Kotrschal et al, Current Biology 23:168, January 21, 2013.)
More about the brain size-gut size problem... Sliced meat: implications for size of human mouth and brain? (March 23, 2016).
Posts on brains include:
* A possible genetic cause for the large human brain (March 25, 2017).
* Mice with human brain cells (April 13, 2013).
* Is it possible that mental retardation could be prevented by a simple prenatal treatment? (January 14, 2013).
Posts on fish include:
* Helicoprion -- a fish with 117 teeth, arranged in a spiral (March 9, 2013).
* Did you see what the sawfish sawed? (April 27, 2012).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
January 23, 2013
Geron sells its stem cell business
January 23, 2013
Briefly noted...
We previously noted that Geron had started a clinical trial of a product based on embryonic stem cells, and that they then pulled out of the stem cell business [links at the end]. The news is now that they have sold the stem cell business. The details are not important for us here, and the deal will not be finalized until late in 2013. But it is good news -- and not surprising -- that the work will continue.
* News story: Geron Sells Stem Cell Assets. (The Scientist, January 8, 2013.)
* Company announcement: Geron to Divest Stem Cell Assets. (Geron, January 7, 2013.)
Background posts:
* Therapy based on embryonic stem cells: the first clinical trial (October 23, 2010).
* Therapy based on embryonic stem cells: the first clinical trial -- follow-up (December 5, 2011).
For more on stem cells: The role of the immune system in making stem cells (February 8, 2013). Induced pluripotent stem cells (iPSC).
Stretchable electric wires
January 22, 2013
Have you ever cut through the insulation of an electrical wire? Did you notice that the metal part -- the inner part that conducts electricity -- drips out onto the floor? No? Well, it would if that inner metallic part were liquid -- as proposed by a group of scientists from the University of North Carolina in a recent article.
Why would you want to do that? Their goal is to make wires that can be stretched. That's not a new idea, but attempts to date have had limited success; ordinary metals just aren't very stretchable. Their approach here is novel. Wire has an outer part, the insulating sheath, and an inner part, the conducting metal. The approach here is to separate the functions of the outer and inner parts more completely. Stretching is the job of the outer sheath; the inner part just conducts electricity. Thus they focus on making a plastic for the sheath that can be stretched. However, the core is a liquid metal. When the sheath stretches (or relaxes), the conducting liquid metal just conforms to the sheath.
Liquid metals? You may know that there is one metal that is liquid (at room temperature), but mercury presumably would not be a good candidate because of toxicity. Perhaps you recall that there is another metal with a melting point just a bit above room temperature; it will melt in your hand. That is gallium. What they do is to dissolve something in the gallium, to lower its melting point. The alloy of gallium and indium that they use has a melting point of 16 °C.
Does their liquid-core wire work? Yes. They provide some lab measurements, and some real use. They use the wire as part of the cable for an earphone or an iPod charger, and show that it works fine, even while the experimenter repeatedly stretches the wire to several times its original length and relaxes it. The movie files listed below capture these exercises.
The following graph illustrates one key feature of their novel wires: the mechanical properties are now due entirely to the outer insulating sheath.
|
The graph shows the stress vs strain relationships for two of their sheath types, made under different conditions. For each, there are two curves. One is for the hollow (empty) sheath(hollow symbols). One is for the sheath filled with its liquid metal conducting core (filled symbols). For each type of sheath, the results with hollow or filled sheath are essentially the same. This shows that they have achieved their goal of separating the roles of the outer and inner parts of the wire.
The y-axis shows the stress: how hard they pulled. The x-axis shows the strain: how much the wire stretched. 100% on the strain scale means that the wire is now twice its original length.
This is Figure 3 from the article.
|
Are there any problems with this liquid-core wire? Yes. If you cut through the insulation, the inner metallic part drips out onto the floor.
News story: Liquid Metal Used to Create Wires That Stretch Eight Times Their Original Length. (Science Daily, December 18, 2012.)
Movies. There are two short videos posted with the article. There is also a video at YouTube, which is equivalent to #2. In the video, someone is stretching and relaxing the earphone cable, but the sound continues normally. Ultra-Stretchable Wires. (YouTube, 37 seconds)
The article: Ultrastretchable Fibers with Metallic Conductivity Using a Liquid Metal Alloy Core. (S Zhu et al, Advanced Functional Materials 23:2308, May 13, 2013.)
A stress-strain curve for a fiber was presented, with more explanation, in the post
Spider silk: Can you teach an old silkworm new tricks? -- Update (February 11, 2012).
More about stretchable electronics: Supercapacitors in the form of stretchable fibers -- suitable for clothing (May 2, 2014).
More about unusual wires: On sharing electrons (May 3, 2011).
More about electrical insulation: Using an electrical conductor as an insulator (September 12, 2021).
More gallium... Capturing carbon dioxide using gallium (April 4, 2022).
A bacterial cocktail to fight Clostridium difficile
January 19, 2013
The bacterium Clostridium difficile (C dif, for short) is becoming a serious medical problem. It is particularly interesting because the C dif problem is not a simple story of an infectious agent, but rather a story of the balance between the microbes of the gut. Many people carry C dif without any problem. However, upon an antibiotic treatment, the C dif may take over and become a major microbe in the gut. It makes toxins, and kills. C dif is not easily treated with antibiotics.
What to do? It would seem that restoring the balance of microbes in the gut would be a good idea. In fact, fecal transplantation, from a healthy donor, is effective in treating C dif.
A recent article reports some interesting work with a mouse model for C dif. The mouse model behaves rather similarly to the human infection. Treatment with feces from a healthy donor is an effective treatment. But then the scientists do something new... they isolate various kinds of bacteria from the feces, and explore the use of cocktails containing defined mixes of specific strains. Turns out, one specific mix works well. This is an interesting and encouraging result.
Here is an example of what they did.
|
In this experiment, they test three bacterial mixes as treatments for a C dif infection in mice. The graph shows the C dif in the feces of the infected mouse (y-axis; log scale) vs time (x-axis). The mice have already been infected; time 0 is the time when treatment occurs.
The y-axis is labeled CFU C difficile/gram feces. CFU means "colony forming units". For those not familiar with the microbiology term, CFU can be considered as the number of bacteria. Thus the y-axis shows number of bacteria per gram of feces of the infected mouse. It is a measure of the infection.
|
At the start (left), you can see that the infected mice are shedding large numbers of C dif -- about 108 per gram feces.
At "Day 0" the infected mice are treated, using three bacterial mixes. For two of them, C dif shedding continues, as before. However, for one bacterial mix (Mix B, green symbols), shedding soon declines, and reaches the detection limit (dotted line), around 102 per gram feces. Importantly, the shedding continues to be negligible -- with only the one treatment at day 0.
This is Figure 4b from the article.
|
The figure shows, then, that a specific mix of bacteria can be used to treat C dif infection in mice. Would this approach work in humans. The only way to know is to try it. Not necessarily the specific bacterial mix that was isolated here, from mice, but the approach.
This should be thought of as more than just exploring treatment for C dif; it is also a step in the broader field of understanding the gut microbiota, and learning how to manipulate it.
News stories:
* Bacterial Cocktail Treats Infection -- Mice fed a mix of six strains of bacteria were able to fight a C. difficile infection that causes deadly diarrhea and is resistant to most types of treatment. (The Scientist, October 29, 2012. Now archived.)
* Fecal Bacteria Overpower Highly Contagious C. diff Strain. (GEN, October 26, 2012. Now archived.)
The article, which is freely available: Targeted Restoration of the Intestinal Microbiota with a Simple, Defined Bacteriotherapy Resolves Relapsing Clostridium difficile Disease in Mice. (T D Lawley et al, PLoS Pathogens 8(10):e1002995, October 25, 2012.)
More about C dif:
* A drug to treat the gut microbiome (February 25, 2023).
* Fecal transplantation as a treatment for Clostridium difficile: progress towards a biochemical explanation (February 8, 2015).
More on the human microbiota...
* Sharing microbes within the family: kids and dogs (May 14, 2013).
* Malnutrition: is more (or better) food the answer? (March 8, 2013).
* Antibiotics and obesity: Is there a causal connection? (October 15, 2012). Gut.
* A virus that could treat acne? (October 21, 2012). Skin.
* Can the Staph solve the Staph problem? (July 12, 2010). Nose -- and an example of exploiting the competition between good and bad microbes.
An easier way to get infected with malaria
January 18, 2013
The normal way one gets infected with malaria is to be bitten by a mosquito that is carrying the parasite, Plasmodium falciparum (Pf). Now, a team of scientists has developed a way to directly inject people with the parasite, avoiding the need for a mosquito.
Why would you want to do that? It's for lab work. One difficulty in studying malaria is starting the infection. The ability to infect a study population directly with a defined dose of parasites will facilitate lab work, including drug testing.
The key part of the work was to develop a well-defined product. The scientists, from a company called Sanaria (get it?), describe it as "aseptic, purified, vialed, cryopreserved Pf sporozoites" (from the abstract; sporozoites are the form of the parasite that mosquitoes inject when they bite you). They now report the results of a small Phase 1 trial of their product.
|
The graph shows the effectiveness of their controlled inoculations. Volunteers were inoculated with defined doses of the sporozoite product. The level of parasites in the blood (y-axis; log scale) was measured over time (x-axis).
Three dose levels of parasite were used, coded by green at the highest dose to red to black at the lowest dose. Five of six volunteers treated at each dose level showed infection. (This is comparable to the infection level achieved using mosquitoes in controlled tests.) The data above are for those who showed infection. (At the end, all volunteers were treated with drugs to eliminate the infection.)
The graph shows that infection proceeded quite regularly in those who achieved infection. The effect of dose is less than expected; this is not understood at this point.
There was little evidence for adverse effects of the treatment, beyond those expected for the infection itself.
This is Figure 1D from the article. Other parts of the figure show the variability between volunteers for each dose.
|
They seem to have achieved their primary goal: the development of a preparation of malaria parasites that can be easily handled, and delivered to people in a controlled manner. This small trial needs follow-up, but it is encouraging. If this holds, the ability to provide controlled infections will facilitate malaria research.
It is also possible that this line of work could lead to a malaria vaccine. There is evidence that inactivated sporozoites could be effective as a vaccine. However, there has been no reliable delivery method. The current work seems to provide that reliable delivery method; this should allow work on a sporozoite-based vaccine to proceed with the ability to do controlled tests. In fact, they have already reported some work on this.
News story: Injectable Formulation of Malaria Parasites Achieve Controlled Infection. (Science Daily, November 13, 2012.)
The article, which is freely available: Controlled Human Malaria Infections by Intradermal Injection of Cryopreserved Plasmodium falciparum Sporozoites. (M Roestenberg et al, American Journal of Tropical Medicine & Hygiene, 88:5, January 2013.)
More on malaria...
* A vaccine against malaria -- with 100% efficacy? (October 20, 2013).
* Malaria-infected mosquitoes have greater attraction for people (May 28, 2013).
* Genes that protect against malaria (January 19, 2010).
And see my page Biotechnology in the News (BITN) -- Other topics under Malaria.
If you would rather read about mosquitoes... Mosquitoes are delectable things to eat (August 21, 2010).
January 16, 2013
Captain Nemo anyone? -- Giant squid sighting
January 15, 2013
Jessica sends (with the title that is the first part of my title above)... http://touch.latimes.com/#story/la-fi-tn-giant-squid-video-20130107/. Elusive giant squid caught on video for the first time. (Los Angeles Times, January 8, 2013.)
(If you have trouble loading this page, just skip it and go on. In fact, the page seems to be no longer available.)
Squid are a bit weird under any circumstances, even when compared with their fairly close relatives, the octopus. The giant squid holds a special place within squid-dom. That's partly because it is indeed a giant, with a body length -- including the tentacles -- approaching that of the large whales. Further, until recently, no one had ever seen one alive. Imagine bizarre 20-meter long (60 feet) creatures known only from specimens found dead on beaches. They are the stuff of legends.
What's new here is the observation of a giant squid in nature -- the first time this strange giant has been seen live in its natural habitat.
Information is very limited at this point. Some videos have appeared, at YouTube and elsewhere. Some have been withdrawn. The Discovery Channel, a co-sponsor of the expedition, is going to present the squid story, on January 27. You might want to watch for it. In any case, more information, and real videos of the main character, will presumably be available after that official roll-out.
Here is a page from Discovery (now Seeker): Giant Squid Filmed in Pacific Depths. (Discovery News, January 7, 2013. Now archived.)
The Figure at the right is a photo of the squid, reduced from one on that page. The animal is about 8 meters (26 feet) long -- about half the size of the largest specimens known.
|

|
Previous Musings squid... Quiz: What is it? (November 20, 2012). See the answer -- and the picture. It's a smaller squid, but gives the idea.
A post about octopus, a close relative of the squid: Why don't your arms get entangled or stuck together? (June 10, 2014).
And then there is... What is it? (December 28, 2010). The squid-worm shown there really has nothing to do with squid -- except the name, chosen to help describe something that makes no sense.
Also see... Sleep stages in octopuses -- do they dream? (July 13, 2021). Includes an extensive list of posts about octopuses and other cephalopods.
Those who don't understand Jessica's title should look up Captain Nemo in Wikipedia: Wikipedia: Captain Nemo. (Are there readers from India who do not know about Captain Nemo? Are there readers from Poland who do not know that Nemo might originally have been Polish?)
For more about squid strandings, have a look at the following news story. It is from a local newspaper, about a squid invasion on northern California beaches last month. These are not giant squid -- merely jumbo. Hundreds of Humboldt squid wash up on Aptos area beaches. (Santa Cruz Sentinel, December 10, 2012.)
Is it possible that mental retardation could be prevented by a simple prenatal treatment?
January 14, 2013
Let's look at some results. We will fill in the background later.
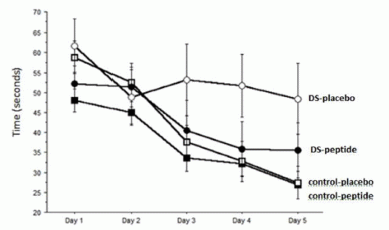
|
The graph shows the results of a learning test for two kinds of mice with and without treatment. The learning score is on the y-axis; lower time is better.
The simplest way to read the graph is to look at the day 5 results (right side). You can see that the lowest scores (best learning) are for the control mice, with either the peptide treatment or the placebo. However, for the DS mice, the scores are very different. The DS mice with placebo had a high score; use of the peptide treatment greatly improved their score.
|
You can also look at the improvement from day 1 to day 5. It's messier, but leads to the same basic conclusion.
The key observation is that the DS-peptide mice do better than the DS-placebo control. It does not matter much whether the DS-peptide mice are as good as the control mice, at least for now.
This is Figure 1 from the article.
|
Something interesting is happening here. What is this about? What are DS mice, and what is this peptide?
DS mice are mice considered a model for Down syndrome (DS), the most common cause of mental retardation in humans. The human DS disease is due to having an extra copy of one chromosome (#21); the DS mice also have an extra copy of a chromosome, one that is partly equivalent to the human 21. We need to emphasize that the precise nature of human DS is not clear, and it is not clear how good the DS mice are as a model of human DS. Nevertheless, DS mice show a learning deficit, as shown here (the DS-placebo results). And the peptide treatment helps.
The "peptide" is actually a pair of small proteins, used together. They are neurotrophic growth factors, known to be involved in brain development. Previous work had suggested they have some benefit for DS mice. What is novel here is the treatment of the mice in utero, leading to an improvement in learning in the adult mice. That is, it would seem that a genetic error affecting brain development can be reversed by a prenatal treatment. That's an interesting finding. Understanding this, and finding whether it is relevant to this -- or any -- human disease requires further work.
News story: Prenatal Intervention Reduces Learning Deficit in Mice. (Science Daily, November 29, 2012.)
The article, which is freely available: Prenatal Treatment Prevents Learning Deficit in Down Syndrome Model. (M Incerti et al, PLoS ONE 7(11):e50724, November 29, 2012.)
More about Down syndrome: Down syndrome: Could we turn off the extra chromosome? (November 15, 2013).
Posts on brains include:
* Fish with bigger brains may be smarter, but ... (January 25, 2013).
* A drug treatment for an autism-like condition in mice (November 9, 2012).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
More about growth factors: Targeting growth factors to where they are needed (April 21, 2014).
Are there really three domains of life?
January 12, 2013
For many years biologists have felt that the best classification of organisms started with dividing them into three domains; the eukaryotes, the archaea and the bacteria. The eukaryotes include animals and plants, plus many microbes (such as the fungi, protists, and true algae). They are characterized by a cell that includes membrane-bounded organelles, such as the nucleus and mitochondria; we call this type of cell a eukaryotic cell. Organisms in the other two domains both have the simpler prokaryotic cell, but archaea and bacteria are distinct in various ways.
The origin of eukaryotic cells is one of the great mysteries of biology. We understand that part of the story involves acquisition of organelles such as the mitochondria and chloroplasts, both of which are clearly derived from bacteria. However, beyond that, the story is unclear. Intriguingly, eukaryotic cells contain features of both archaea and bacteria.
In fact, there is an alternative to this common three-domain model. It is introduced in the following figure. The figure shows a pair of trees showing two possible relationships between major groups of organisms.
|
In each tree, time goes from left to right. An ancestor, unspecified and unlabelled, is at the left. Eukaryotes and bacteria are shown as single groups (green and brown triangles, respectively). Various subgroups of archaea are shown, because they are relevant to the story. All the archaea are shown with a gray background.
Lest we get lost in all the big words there, the difference between the two trees is that the archaea are all together in one tree (left, one big gray box), and are split in the other tree (right, two smaller gray boxes). Another way to look at this is that the order of the top two items, the Euryarchaeota and the Eukaryota, is switched.
This figure is from the Zimmer news story. It is attributed there to the lead author of the article.
|
So what is the big deal? Look carefully at the tree on the left. Look at the first two splits (branchings). At that point we have three groups: the bacteria (bottom), the eukaryotes (top) and the archaea (middle, the gray box). Now look at the tree on the right. After two splits, we have the bacteria as one group, as before. But the other two groups are both sub-groups of the archaea -- and the eukaryotes are down there inside one of those sub-groups of archaea.
That is, in one tree the eukaryotes are a sister group to the archaea; this is the "conventional" three-domain model. In the other tree the eukaryotes arise from within the archaea; this is not consistent with that "conventional" three-domain model. Now, the right-hand tree (sometimes called the eocyte model) has been around for a while, but the reason this comes up now is that a new article reports what they suggest is the best test ever of these two models. Their results support the right-hand tree -- the eocyte model, with eukaryotes arising from within the archaea.
Is this important? As noted above, the origin of the eukaryotes is one of the great mysteries of biology. Now we see that our usual starting point, the three-domain model, is being seriously challenged. Perhaps we know even less about the origin of eukaryotes than we thought we did. This is a story that will long continue.
News story: Redrawing the Tree of Life. (Carl Zimmer, The Loom (National Geographic blog), December 20, 2012.)
The article, which is freely available: A congruent phylogenomic signal places eukaryotes within the Archaea. (T A Williams et al, Proc. R. Soc. B 279:4870, December 22, 2012.)
The post immediately below is closely related to this post. Carl Woese and the archaea (January 12, 2013)
More on the origin of eukaryotic cells:
* An Asgard in culture (February 4, 2020).
* Our Loki ancestor? A possible missing link between prokaryotic and eukaryotic cells? (July 6, 2015). This is very much a follow-up to the current post.
* Origin of eukaryotic cells: a new hypothesis (February 24, 2015).
* Tarsier; eukaryotic cells (August 31, 2009)
Carl Woese and the archaea
January 12, 2013
A historic paper -- from just 35 years ago. It is Carl Woese's paper declaring what has come to be known as the three-domain model for classifying organisms. It's not only a paper of historic significance, but it has long been recognized as such.
There are a couple of reasons to take note of this classic paper here.
* An accompanying post, immediately above, offers a challenge to Woese's three-domain model.
* Carl Woese died recently, on December 30, 2012, at age 84.
What Woese did in 1977 was to compare the gene sequence for a particular gene (for the small ribosomal RNA) for a range of organisms. This was an early example of using gene sequence to determine relatedness of organisms. The key result was that the genes from one group of organisms, which had been considered bacteria, were as different from the other bacteria as from the eukaryotes. This group of organisms was the methanogens. Woese suggested, then, that there are three "equal" groups -- or domains -- of organisms: the bacteria, the archaea, and the eukaryotes (using modern terminology).
So we have a new article challenging the Woese model coming out more or less as he dies. Does the new work diminish the importance of Woese's work? Hardly. We can break down Woese's contribution into three parts.
* He established an experimental approach for measuring relatedness, using genome sequence.
* He established the distinction between the archaea and the bacteria. That is, he recognized the archaea as a distinct group. (Back in 1977, he called them "archaebacteria".)
* He proposed the three-domain model.
The first two stand. Woese introduced using genome sequence for phylogenies, and he brought us the archaea. The third dominated discussion for many years, but seems open to question. Woese had few archaea in those days. The new work makes use of much more information on the archaea, and primarily deals with the relationship between the archaea and the eukaryotes. We do not yet know if the new work will stand; some think it makes the best case yet for the alternative, eocyte, model. If the eocyte model wins, it can be thought of as building on what Woese did. Woese introduced us to the archaea; they are more complex than he knew at the time.
News story: Carl R. Woese, who discovered a new domain of life, dies at 84. (University of Illinois, December 31, 2012.)
The 1977 article is recognized by the journal that published it as a PNAS classic. The first two articles listed here are PNAS items, from 2012, about the original article. The "Profile" is a brief overview; the "Perspective" is a more detailed scientific analysis. Both contain modern versions of the three-domain model. The third here is the original article. All are freely available.
* Profile: Woese and Fox: Life, rearranged. (P Nair, PNAS 109:1019, January 24, 2012.)
* Perspective: Phylogeny and beyond: Scientific, historical, and conceptual significance of the first tree of life. (N R Pace et al, PNAS 109:1011, January 24, 2012.)
* The article: Phylogenetic structure of the prokaryotic domain: The primary kingdoms. (C R Woese & G E Fox, PNAS 74:5088, November 1977.) It's a short article that remains quite readable to this day. Those with some background in microbiology, or at least biology, may enjoy looking over the original article.
Related accompanying post (immediately above): DNA: Are there really three domains of life? (January 12, 2013).
And more...
* The Asgard superphylum: More progress toward understanding the origin of the eukaryotic cell (February 6, 2017).
* Our Loki ancestor? A possible missing link between prokaryotic and eukaryotic cells? (July 6, 2015).
More about the domains of life... The largest known virus (August 5, 2013).
More about methanogenic archaea: What caused the mass extinction 252 million years ago? Methane-producing microbes? (October 12, 2014).
More about ribosomal RNA: Ribosomes with subunits that are tethered together (October 5, 2015).
* Previous history post... DNA: Watching the hopping supercoils (November 24, 2012). (This is not primarily a history post, but includes some history in the note at the end.)
* Next... The Mudville story, on its 125th anniversary (June 3, 2013).
My page Internet resources: Miscellaneous contains a section on Science: history. It includes a list of related Musings posts.
Book on the Archaea... See my page Books: Suggestions for general science reading: Forterre, Microbes from Hell (2016).
January 9, 2013
The placebo guy
January 9, 2013
We recently noted an article about a gene that affects a placebo effect [link at end]. One of the lead authors of that study was Dr Ted Kaptchuk, of Harvard. The university news magazine has just published a news feature about Kaptchuk. It is an interesting overview of the field and the person. I encourage you to look over this news item. (It may even make you feel better.)
News story: The Placebo Phenomenon -- An ingenious researcher finds the real ingredients of "fake" medicine. (C Feinberg, Harvard Magazine, January-February 2013, p 36.)
Background post: The placebo effect: a mutation that makes some people more likely to respond (October 30, 2012).
More from the same lab... Would a placebo work even if you knew? (January 31, 2014).
Making an artificial ion channel from DNA
January 8, 2013
There are various reasons why a story catches my attention and gets considered for a Musings post. Striking pictures are one way to get my attention.

|
At the left is a striking figure. It's a diagram, of course, but the scientists followed through and made what they planned, so let's look.
What is it? An ion channel, or "nanopore". It's a complex structure that can insert itself into a lipid membrane, where it will serve as a channel through the membrane.
This is reduced from the first figure in the Phys.org news story. It is probably the same as part of Figure 1A from the article.
|
Ion channels are common -- and important -- in biology. Ion channels in the cell membrane allow common ions, such as Na+, to get in or out of the cell. Such ions are the basis of voltage differences across cell membranes, and ion channels are central to the operation of the nervous system. What makes this one special is that they make it almost entirely from DNA. The idea is to design strands of DNA that will self-assemble into the desired structure, the ion channel in this case. (Just to be absolutely clear... We are not talking here about DNA coding for the channel; the DNA is actually the construction material for making the channel.)
|
And here it is. Actually, several of them, all attached to a small blob in the lab. The surface of the blob, or vesicle, is a lipid membrane, much like that of biological cells.
This is reduced from the second figure in the Phys.org news story. It is probably the same as Figure 1F from the article.
|
They do an interesting "trick" to help get the channel device to stick to the membrane. Look at the diagram (the upper picture). The small orange (carrot-like) things? They are modified DNA structures with cholesterol attached. The cholesterol helps to hold the device to the membrane. Cholesterol is not part of natural DNA; here, it is attached to some of the DNA bases. The heart of the construction process involves making use of DNA base pairings, but it is fine to have other structures attached. (The red part at the bottom of the diagram? That is the part that goes through the membrane. The set of red cylinders forms a pore in the middle.)
Importantly, they test the blobs-with-DNA-based-channels; they work! The scientists observe a voltage across the membrane when the channel device is inserted.
A striking picture got my attention. But there is more here than just a pretty picture. The work involves constructions with DNA -- not just to make a structure, but to make a functional device. This is probably one of the most complex DNA constructions done to date.
News story: Artificial ion channels created using DNA origami. (Phys.org, November 16, 2012.)
* News story accompanying the article: Chemistry: Functional DNA Origami Devices. (M S Strano, Science 338:890, November 16, 2012.)
* The article: Synthetic Lipid Membrane Channels Formed by Designed DNA Nanostructures. (M Langecker et al, Science 338:932, November 16, 2012.)
A recent post on "DNA technologies": Making big "molecules" from big "atoms" (December 7, 2012).
Nanopores -- another revolution in DNA sequencing? (June 22, 2012). The use of the channels in this new work is somewhat similar to how channels are used in the new method of sequencing DNA using nanopores. (In fact, the protein used in that method by some groups was one of the models they used here to guide their work.)
Here are some posts that note the function of natural ion channels. One discusses a disease related to an ion channel.
* A drug treatment for an autism-like condition in mice (November 9, 2012).
* How an octopus adapts to the cold -- by RNA editing (March 5, 2012).
* How to find the blood (August 29, 2011).
A computer model of a cell
January 7, 2013
An interesting development... Researchers at Stanford University have developed a model of a cell. Not a plastic model showing you the main parts, but a detailed computer model, simulating everything going on in the cell. In effect, they have made a version of the cell in silico.
The cell they chose to model was one of the very simplest cells known, the tiny bacterium Mycoplasma genitalium. Its genome contains only 525 genes -- about 1/10 as many as the common bacterium Escherichia coli. They based the workings of their model on all the data they could find.
|
An overview of the model. At the left is a list of 16 "Cell variables" that they calculate to characterize the cell. At the right is a list of 28 "Cell process submodels", which are the parts of their computer program. The lines connecting the two sides show which variables feed into which submodels.
After each iteration of the model they ask if the cell has divided; see the far right. If so, the model is terminated.
This is Figure 1B from the article.
|
What is the point? Perhaps most importantly, it is a test of how well we understand the cell. Does the model behave like the cell, or does it say things that we know are wrong?
How good is the model? One simple test they did was to see if each gene was "essential". That is, they deleted each of the 525 genes, one at a time. Did the strain -- the computer model of the strain -- still grow with that gene gone? This experiment has actually been done with real bacteria, so they could compare the results. The computer model was about 80% accurate. Its errors were in both directions: predicting that a gene is essential when it is known to be non-essential, and vice versa. Judging that result as good or bad would miss the point. They will now examine each error, and see if they can figure out why the model was wrong. That leads to improvement of the model.
As people gain confidence in the model, they may use it to predict new things. They can then test the predictions. The tests feed back to improve the model, as necessary. In the long run, that is the point. A computer model such as this embodies our understanding -- and is a tool in helping us improve our understanding. The work reported here is a tremendous achievement -- the best such model yet. Understanding its mistakes is a key part of improving the model and -- therefore -- of improving our understanding of the biology.
News story: Researchers Produce First Complete Computer Model of an Organism. (Science Daily, July 21, 2012.)
* News story in Nature: Systems biology: A cell in a computer. (M Isalan, Nature 488:40, August 2, 2012.)
* The article: A Whole-Cell Computational Model Predicts Phenotype from Genotype. (J R Karr et al, Cell 150:389, July 20, 2012.)
Thanks to Thien for originally alerting me to this work -- back when it was new. It took me a while to figure out what to do with it. The article itself is not easy reading, but it does represent an interesting development that is worth briefly noting.
More about computers: Alan Turing -- and the music of Iamus (November 14, 2012).
How porcupine quills work
January 5, 2013
The picture above is the tip of a quill from a North American porcupine, as observed by scanning electron microscopy (SEM). The amount of tip shown in the picture is about a half millimeter. Note that the tip is not smooth, but rather has flap-like features -- called barbs.
The picture of the quill is trimmed and reduced from one in Ed Yong's story listed below. Similar SEM figures in the paper show the barbs. The picture linked above is also from that story. (You did see a face in there? If not, go back and look again.)
So what brought this up? A team of scientists, largely from Harvard and MIT, has studied the forces required to insert and retract quills, as well as quills that have had their barbs removed. They then designed artificial porcupine quills, using what they learned from studying the real thing. They suggest that what they learned may be useful in designing improved medical devices such as needles or sutures.
Perhaps their key discovery is that barbed quills penetrate the skin more easily than de-barbed ones -- even though the barbed quills are larger (in diameter). This is probably because the uneven surface of the barbed quills penetrates mainly at the outermost part. Thus the force of penetration is focused onto a small region of the quill. Of course, as is well known, removal of barbed quills is difficult.
News stories:
* Inspiration from a Porcupine's Quills. (Science Daily, December 10, 2012.)
* Why porcupine quills slide in with ease but come out with difficulty. (E Yong, Not Exactly Rocket Science (Discover blog), December 11, 2012.)
The article: Microstructured barbs on the North American porcupine quill enable easy tissue penetration and difficult removal. (W K Cho et al, PNAS 109:21289, December 26, 2012.)
A previous post about better needles for inoculations... A better way to deliver a vaccine? (July 25, 2010).
And sutures and such...
* Fixing the heart with some glue and light (July 27, 2014).
* Smart sutures (November 3, 2012).
This work is, in part, an example of bio-inspired (or bio-mimetic) development. For more about this emerging field, see my Biotechnology in the News (BITN) topic Bio-inspiration (biomimetics). It includes a listing of some other Musings posts in the area.
One example... Bending a rigid rod (May 17, 2013).
January 3, 2013
A simpler assay for detecting low levels of HIV, using gold nanoparticles
January 3, 2013
Let's jump in and look at a key result...
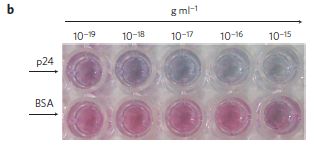
|
This shows a test for a protein called p24, which is part of the human immunodeficiency virus (HIV).
From left to right, each well contains more protein, as shown by the numbers across the top. The top row contains wells with the HIV p24 protein; the bottom row has a control protein, BSA (bovine serum albumin).
|
All the control wells (BSA) are red. For p24, most are blue, at least past the very lowest level.
This is part of Figure 4b from the article.
|
That is, this assay can detect very low levels of this HIV protein -- and it can be read "by eye", without needing a machine. And that's the point of the scientists who recently reported this novel assay. Let's look more closely at how they did it.
Here is their diagram for the key part of this assay.
|
They use a solution of gold ions, Au3+. The gold ions get reduced to gold atoms, Au (or Au0). Depending on exactly how this happens, they get red or blue particles of gold (as shown at the right). These red or blue gold particles are what you saw in the first figure, at the top.
What determines whether we get red or blue particles? The type of particle is controlled by the hydrogen peroxide (H2O2), shown in the reaction equation (part a of the figure; left). If there is plenty of the H2O2, they get red (part b -- the upper branch). But if the H2O2 is depleted, they get blue. How does that happen? It's part of the assay design. If the protein (p24 in this case) is present, the antibody recognizes it, and eventually captures an enzyme (catalase) that removes the H2O2. This is shown in part c -- the lower branch of the figure.
In summary...
* If the protein being tested for is absent, the peroxide leads to red gold particles.
* If the protein is present, catalase destroys the peroxide, and they get blue gold particles.
This is Figure 2 from the article.
|
This is clever. They have achieved some of their goals. The assay is quite sensitive, and it does not require a machine to read the result. Is the assay useful at this point? I'm a little skeptical. It is still complex, and requires reagents that need to be kept refrigerated (the antibodies and enzymes). It doesn't seem ready for work in the remote field. However, it is a useful step, and it will be interesting to see what further developments occur.
The assay here is a variation of the ELISA, which stands for enzyme-linked immunosorbent assay. ELISAs use an antibody to capture a specific protein, and then couple this to an enzyme reaction that produces a measurable product. The novelty of the new work is the type of measurable product. This is made possible by their understanding of gold nanoparticles, and then by finding a role for an enzyme in developing those particles.
News stories:
* Test Developed to Detect Early-Stage Diseases With Naked Eye: Prototype Ultra Sensitive Disease Sensor Developed. (Science Daily, October 28, 2012.)
* Naked Eye ELISA Developed as Biomarker Diagnostic. (GEN, October 29, 2012.)
The article: Plasmonic ELISA for the ultrasensitive detection of disease biomarkers with the naked eye. (R de la Rica & M M Stevens, Nature Nanotechnology 7:821, December 2012.)
More on HIV...
* Infant cured of HIV? (April 15, 2013).
* How a drug can cause an autoimmune reaction (September 1, 2012).
More hydrogen peroxide:
* Electronic monitoring of plant health; it might even allow an injured plant to call a doctor (June 21, 2020).
* Self-powered micromotors for speeding up chemical reactions, such as destruction of chemical weapons (March 14, 2014).
More gold nanoparticles: Air pollution: progress towards a process for ammonia oxidation (April 5, 2019).
More gold... Prospecting for gold -- with help from the little ones (March 1, 2013).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on HIV
Crocodilians: Thick-skinned but very touch-sensitive
January 2, 2013

|
A crocodile head.
What's important here are the many tiny bumps over much of the surface. These bumps are called integumentary sensory organs (ISOs).
There are about 3,000 ISOs around the head region of the crocodile -- and more on the rest of the body. Each ISO is about 1 millimeter across.
This is Figure 2C from the article.
|
People have known about the ISOs for a while, and known that they are sensory organs. However, their biological function has been unclear. A new article reports more in-depth work on the ISOs of crocodilians (alligators and crocodiles). Among the findings... crocodilian ISOs are more sensitive to the touch of a hair than are human fingertips.
The paper includes anatomical work, examining the structure of the ISOs and their connection to the nervous system. It also includes functional tests, such as the one noted above.
What are the ISOs for? Think about the situation... an armor-plated animal -- with ultra-sensitive mechanosensory sites. They can detect small water ripples, such as those caused by possible prey. And they can detect contact with prey and whatever else is around. It seems likely that the high density of the ISOs around (and in!) the mouth aid is distinguishing food from debris -- and from the kids. Female alligators gently carry their young in their jaws.
News story: Despite Their Thick Skins, Alligators and Crocodiles Are Surprisingly Touchy. (Science Daily, November 8, 2012.)
* News story accompanying the article: Croc jaws more sensitive than human fingertips. (K Knight, Journal of Experimental Biology 215(23):i, December 1, 2012.)
* The article: Structure, innervation and response properties of integumentary sensory organs in crocodilians. (D B Leitch & K C Catania, Experimental Biology 215:4217, December 1, 2012.) There is a 3-minute movie file associated with the article. It is freely available from the article web site; choose "Supplementary Material". That takes you to a description of the various scenes, and a link to the movie file. Some scenes include a human hand, which will give you a size comparison.
If you find yourself paying more attention to parts of the structure other than the little bumps, you should know that the work here used juveniles, only a few centimeters long. As noted above, the movie file with the article lets you compare the size of the animals with a human hand.
More on the crocodilians:
* Crocodiles, humans, and stones (November 13, 2009).
* Space shuttle: some final photos (December 3, 2012). You'll have to look around a bit.
More about the sense of touch... eSkin: Developing better sense of touch for artificial skin (November 29, 2010).
Top of page
The main page for current items is Musings.
The first archive page is Musings Archive.
E-mail announcements -- information and sign-up: e-mail announcements. The list is being used only occasionally.
Contact information
Site home page
Last update: February 25, 2026