Home
> BITN
> DNA and the genome
| Contact
Biotechnology in the News (BITN)
DNA and the genome
Introduction
Overview
An introduction to DNA: basic structure and how it replicates
Commemorations of the 50th anniversary of the Watson-Crick DNA structure
The human genome
Examples of how genome information is useful
Web sites (general)
Books
Posts in my Musings newsletter. Also see next section.
Miscellaneous articles (2007-2010). Also see previous section.
|
|
Links to external sites will open in a new window.
|
|
Bottom of page; return links and contact information
|
Introduction
A major news story over recent years has been the announcement of the genome sequence for humans. In fact, this project reached a symbolic completion point in April 2003. But this human genome work is just part of a much bigger story -- which includes a list of many completed genomes, for microbes, plants and animals. And all this genome work is just the beginning... genome information alone does not solve anything in particular; it is a big resource that will make further biological work easier.
top of page
Overview
This topic was discussed in the BITN class, Fall 2003. This overview section summarizes the class presentation. The original web materials were designed as a supplement to that class presentation.
Two major news stories of 2003 set the background for this discussion. One is the 50th anniversary of the announcement of the double helical structure of DNA. The other is the announcement of the completed DNA sequence for the human genome. We discussed the development of the DNA structure. A key idea that emerged from this is the complementarity of the two DNA strands. This complementarity immediately suggests how DNA replicates -- by the two strands separating and each serving as a template for a new strand. The resulting "daughter" DNA molecules have one "old" strand and one "new" strand. A physical test of replicated DNA, showing this characteristic, was key in "proving" the basic DNA model. There is much chemical complexity to DNA and much biochemical complexity to how DNA really replicates, but the basic logic of a double stranded structure held together by complementarity still holds.
We then discussed DNA sequencing. We started by looking at some simple DNA sequencing results -- and showed how easy it is to actually read the sequence. Of course, what we looked at is the end step of a lengthy series of steps. We discussed an example of how one might generate the pattern we saw on the sequencing film; our example was not what is actually done, but was a simpler variation to illustrate the logic. The main problem with this basic sequencing procedure is that it works for only about 500 bases. Thus sequencing larger genomes requires some additional work, but it is still based on the same classical procedure that we started with. For large genomes, the process is highly automated, including the use of lasers to read dye-coded bases. Further, tremendous computer capability is needed to keep track of the data from the millions of pieces of DNA that are individually sequenced.
We discussed the gene count for humans. It is rather low -- and also uncertain. It is uncertain because we actually have considerable difficulty recognizing genes simply from DNA sequences, especially for complex organisms. The low gene count is forcing us to emphasize complexities in gene function, such as splicing and editing, that allow more than one protein to be made from a gene. We then discussed applications of genome information, especially of genome differences between individuals. These include applications such as forensic testing and paternity testing, which were developed some time ago. We discussed some drugs which are chosen based on specific genetic characteristics -- either of the individual, or even of the particular cancer. We then discussed more recent work, using gene chips (microarrays), where analysis of many genes allows leukemia (or leprosy) sub-types to be recognized. The specific figure that I showed was from a recent supplement to The Scientist: New Frontiers in Cancer Research, Sept 22, 2003. One topic that came up during general discussion was prions; I now have a page on prions.
top of page
An introduction to DNA: basic structure and how it replicates
The human genome is made of DNA -- as is the genome of almost all organisms. (A few viruses use the closely related chemical, RNA, for their genome; RNA operates by the same basic principles as DNA in this role.) A major milestone in the history of DNA is being celebrated in 2003 (the year this page was started)... It was fifty years ago, April 1953, that Watson & Crick announced that they had determined the structure of DNA -- a structure that in fact "made clear" how it works.
|
|
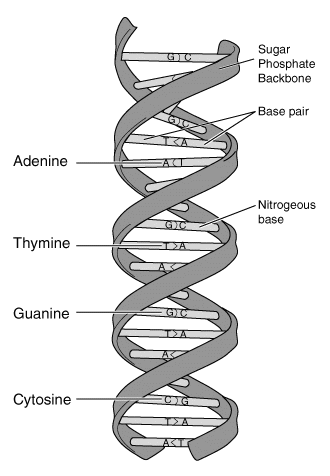
The Fig at the left is a diagram of the general structure of DNA. It shows the famous overall double helix. And it shows the four bases (A, T, G and C) -- which are the "information". At each rung along the DNA ladder is a base pair. Each pair is either A with T or G with C; that is, one strand precisely determines the other strand -- and that indeed is the key to how DNA replicates. See next Fig.
This Fig is from the Glossary of the NIH genome site, https://www.genome.gov/genetics-glossary. Choose deoxyribonucleic acid (DNA). Also see next Fig. (The figures I have used from that site may no longer be there; some have been replaced by newer figures.)
|
|
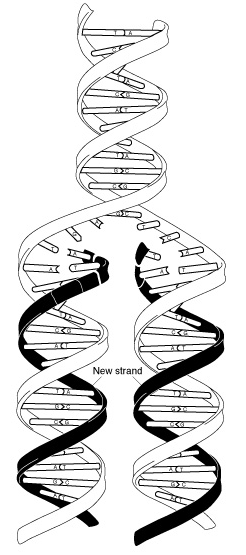
The Fig at the left is a diagram of DNA replicating. The top of the Fig shows a "parental" DNA molecule; the bottom shows two "daughters". During DNA replication, the two parental strands separate, and each serves as the template for a new strand, which is made by those simple base pairing rules (A-T and G-C), which were mentioned with the Fig above.
In this Fig, the "replication fork" (the site and apparatus for making new DNA) is moving upward.
This Fig is also from the Glossary of the NIH genome site, https://www.genome.gov/genetics-glossary. Choose DNA replication.
|
Good overview of DNA, by David Goodsell. This is a "Molecule of the Month" feature at the Protein Data Bank. https://pdb101.rcsb.org/motm/. This is from the educational resources of The Nucleic Acid Database Project at Rutgers.
top of page
Commemorations of the 50th anniversary of the Watson-Crick DNA structure
The double helix structure was published by Watson and Crick in 1953 in the journal Nature. 2003 is the 50th anniversary of that landmark, and there are many commemorations. The January 23, 2003, issue of Nature has a big feature on this. It includes an introductory article (Nature 421:310), copies of the original papers on DNA structure, and many articles discussing various aspects of the DNA story. And then there is more in the April 24, 2003, issue. This includes an article (Nature 422:835) by Francis Collins et al on the future of the human genome project. Fig 1 of that article is a fold-out timeline "Landmarks in Genetics and Genomics"; this is available as a pdf file from the Nature web site. At least some of this material could be usefully read or browsed by those with little background in the field.
Links to items mentioned above:
* Introductory article in the January 23, 2003, issue of Nature (Nature 421:310): https://www.nature.com/articles/421310a. This article is not freely available. You can click on "Table of Contents" for the issue listings; there is no way to tell what is freely available except to try it.
* The Collins article: https://www.nature.com/articles/nature01626. This article is freely available.
* The Table of Contents for that issue: https://www.nature.com/nature/journal/v422/n6934/index.html. Again, there is no way to tell what is freely available except to try it.
More at the Nature site:
* The Nature "web focus" Double helix: 50 years of DNA... https://www.nature.com/nature/dna50/index.html.
* A Nature News Special on the DNA Anniversary... https://www.nature.com/news/specials/dna50/index.html.
Among other web sites that resulted from the commemoration of the DNA anniversary...
* The Cold Spring Harbor Laboratory (long headed by Watson) proclaimed itself the Official Site of the 50th Anniversary of the DNA Double Helix. They no longer maintain the anniversary celebration page, but they have much about DNA... Go to the Dolan DNA Learning Center: https://www.dnalc.org/websites/. Sections of interest there include DNA from the Beginning and DNA Interactive -- and more (from "websites" list at the left). Also, choose the "Resources" section (top menu) and find The Biology Animations Library, with DNA methods such as PCR and Southern blotting. DNA from the Beginning is also available in Chinese, Danish, French, Icelandic, Italian, Portuguese (with German, Spanish promised soon); it is also listed for Molecular Biology Resources: Methods. Another section, Inside Cancer, is listed for BITN Resources: Miscellaneous -- Cancer.
* And from our local Exploratorium: https://www.exploratorium.edu/origins/. Choose Cold Spring - DNA.
top of page
The human genome
There are numerous posts in my Musings newsletter on genome issues, including the human genome. They are listed below in the section Posts in my Musings newsletter.
The human genome was officially announced in February 2001 by two groups.
* The work of Celera Genomics, headed by Craig Venter, was published in Science for February 16, 2001. That entire issue is available online, with free access, at https://www.science.org/toc/science/291/5507.
* The work of the public Human Genome Project, headed by Francis Collins (and earlier by Watson), was published in Nature for February 15, 2001: https://www.nature.com/nature/volumes/409/issues/6822.
The main genome articles are probably too technical for most, but the issues contain many news stories dealing with various aspects of the project.
Nature: Human Genome Collection. https://www.nature.com/nature/supplements/collections/humangenome/index.html. Links to all human genome work from Nature journals. Much consists of the technical articles, but there are also news stories and discussions.
Neandertal genome. February 2009 brings the announcement of a genome sequence from a 38,000 year old Neandertal. It is actually fairly rough at this point, but it is a remarkable achievement to get this far. There is little to conclude for now, except that the genome evidence so far provides no evidence for interbreeding between Neandertals and modern man (Homo sapiens).
* A news story on this announcement, which was made at the AAAS annual meeting: First draft of Neanderthal genome is unveiled; February 12, 2009. https://www.newscientist.com/article/dn16587-first-draft-of-neanderthal-genome-is-unveiled/.
* An excellent longer story from Science. It includes basic background information on Neandertals, and on the genome project. Neandertal Genomics: Tales of a Prehistoric Human Genome. Science 323:866, 2/12/09. It is online at:. https://www.science.org/doi/full/10.1126/science.323.5916.866.
Genome results are so important and fascinating that rodents have been seen scrutinizing their genome data. http://news.bbc.co.uk/2/hi/science/nature/424076.stm. (My main purpose in giving this link is for the Figure, for fun. But the work described there is an example of moving a gene from one organism to another, and using that as a tool to learn about the characteristics of an organism.)
top of page
Examples of how genome information is useful
As noted earlier, the genome is just data. It is not the magic solution to anything in particular. Because the genome data is fairly new, in fact few practical advances can be directly attributed to it. So, much of what I do here is to show how genome info might be used.
Pharmacogenomics and nutrigenomics. Traditional recommendations about proper nutrition and medicine assume that the population is uniform. Data is collected about population averages and this is used to guide medical treatments and nutritional advice. But we are not all the same. In fact, some examples of genetic differences in how we respond to drugs or nutrients have been found, more or less accidentally, in the past. The availability of complete genome information will allow such knowledge to come more rapidly. Briefly, pharmacogenomics is the customization of drug usage depending on an individual's genetic makeup; nutrigenomics is the analogous customization of nutrition information depending on an individual's genetic makeup.
Here is a major nutrigenomics site:
* The European Nutrigenomics Organisation (NuGO): https://www.nugo.org.
That site is also listed on my page Further reading: Medical topics, under Web Sites.
The Future of Nutrigenomics - From the Lab to the Dining Room. A brochure for the general public, from the Institute for the Future. March 2005. https://www.iftf.org/our-work/global-landscape/global-food-outlook/the-future-of-nutrigenomics/.
Cancer. Two articles on work to classify cancers by gene expression patterns. This work has implications for customizing treatment. A Gianella-Borradori et al, Reducing risks, maximizing impact with cancer biomarkers and B A Maher, The makings of a microarray prognosis. The Scientist March 15, 2004, pp 8 & 32. Both are freely available online: https://www.the-scientist.com/opinion-old/reducing-risks-maximizing-impact-with-cancer-biomarkers-50372 and https://www.the-scientist.com/hot-paper/the-makings-of-a-microarray-prognosis-50349.
Race. Is "race" a useful criterion for guiding medical treatment? The important point for us here is that genomics is offering new insight into this socially-charged question. At this point, genetic analysis suggests that there are some genes that reflect "geographical origin", but that the variability of human genomes within any "race" is far more than the genetic differences between "races". Of course, this information will be of more practical use as details emerge.
The following New York Times article discusses a clinical trial of a drug that is being targeted to and tested with only one racial group -- with the approval of the FDA. U.S. to Review Heart Drug Intended for One Race, June 2005. https://www.nytimes.com/2005/06/13/business/us-to-review-heart-drug-intended-for-one-race.html.
The following two short essays are by scientists discussing the race issue:
* M W Feldman et al, A genetic melting-pot. Nature 424:374, 7/24/03. A "Concept essay". https://www.nature.com/articles/424374a.
* S B Haga & J C Venter, Genetics: FDA races in wrong direction. Science 301:466, 7/25/03. "Policy forum". This article explicitly addresses -- and questions -- FDA guidelines for collecting racial data in clinical trials. https://www.science.org/doi/full/10.1126/science.1087004.
Personalized medicine. There are now companies that will take your DNA (and some money) and report back to you your risk for certain diseases. A good idea in principle, but how good is it in practice. Genome pioneer Craig Venter and colleagues have evaluated a couple of these companies, and offer some suggestions. As a general perspective, they think the companies are doing high quality work, technically, but the quality and usefulness of the information is questionable. It is true that your DNA contains information about disease susceptibility, but current knowledge of that is limited -- more limited than the companies want to admit. The paper is: P C Ng et al, An agenda for personalized medicine. Nature 461:724, October 8, 2009. https://www.nature.com/articles/461724a. (If you put the title into Google Scholar, you may find a copy of the paper that is freely available.)
There are many Musings posts in the broad area of personalized medicine. One of the first was: Personalized medicine: Getting your genes checked (October 27, 2009). It links to several others in the area.
top of page
Web sites (general)
This section and the following one used to be together. They are now separated.
Genetics Home Reference, an educational site on genetic diseases in humans; from the National Library of Medicine. It is now part of the Medline Plus site. https://medlineplus.gov/genetics/.
top of page
Books
This section and the one above used to be together. They are now separated.
Books listed here are included on my page Books: Suggestions for general science reading. For each book, there is a link to it on that page for more information.
Books are listed here in date order, most recent first.
David Reich, Who We Are and How We Got Here -- Ancient DNA and the new science of the human past. Pantheon, 2018. ISBN 978-1101870327. See my Book suggestions page: Reich (2018).
Svante Pääbo, Neanderthal Man -- In search of lost genomes. Basic Books, 2014. ISBN 978-0-465-02083-6. A book about ancient human genomes, by the scientist who started the field with the genomes of a Neanderthal and a Denisovan. See my Book suggestions page: Pääbo (2014).
James D Watson (with Andrew Berry), DNA - The Secret of Life. Knopf, 2003. See my Book suggestions page: Watson (2003).
Brenda Maddox, Rosalind Franklin - The dark lady of DNA. Harper/Collins, 2002.
See my Book suggestions page: Maddox (2002).
There is a short essay about Franklin, in the general spirit of the book, online in the Mill Hill collection: K Rittinger & A Pastore, Rosalind Franklin - The dark lady of DNA: https://web.archive.org/web/20170405005602/http://www.historyofnimr.org.uk:80/mill-hill-essays/essays-yearly-volumes/2002-2/rosalind-franklin-the-dark-lady-of-dna/. (Now archived.)
top of page
Posts in my Musings newsletter
Also see next section.
There are many Musings posts on genome and sequencing issues. Each listed here links to related posts. Posts about RNA genomes are included here.
* One was noted above, in the section on "Examples of how genome information is useful": Personalized medicine: Getting your genes checked (October 27, 2009). This includes an extensive list of related posts; most of these are not included below.
More...
* Beethoven's genome (June 26, 2023).
* Briefly noted... Human origins -- more complexity (June 21, 2023).
* The nanobacteria of Omnitrophota (June 5, 2023).
* Briefly noted... New genes -- which have arisen "recently" in the human lineage (May 17, 2023).
* A gene for insomnia? (February 4, 2023).
* Briefly noted... The oldest known sequenced DNA: another new record (December 13, 2022).
* Briefly noted... Why some species of rockfish live for 200 years (April 6, 2022).
* What caused the extinction of the mammoths -- I (January 16, 2022).
* Briefly noted... Ebola transmission from long-term survivors? (October 20, 2021).
* Project Baby Bear: Is it worthwhile to sequence the genomes of babies with mysterious ailments? (October 18, 2021).
* Briefly noted... The oldest known genome: a new record (August 31, 2021).
* Effect of Chernobyl exposure on the rate of new mutations in the next generation (June 1, 2021).
* Nanopore sequencing of DNA using synthetic membranes (May 15, 2021).
* Briefly noted... A new record: the largest known animal genome (March 31, 2021).
* Giraffe physiology: studying a giraffe gene in mice (March 28, 2021).
* Susceptibility to severe COVID: role of a genetic region from Neandertals (February 20, 2021).
* The oldest known identical twins (December 7, 2020).
* Why are some people "elite controllers" of HIV? (December 1, 2020).
* The basis of intersexuality in moles (November 28, 2020).
* A claim of finding dinosaur DNA (May 31, 2020).
* Spike D614G: An emerging mutant strain of the virus that causes COVID-19 (May 12, 2020).
* The microbiome of babies born by C-section (November 2, 2019).
* Domestication of the almond (August 26, 2019).
* The genetic basis of why parrots seem so human (March 31, 2019).
* Reconstructing an ancient enzyme (February 26, 2019).
* Identifying people from public DNA databases -- even if they are not included (January 20, 2019).
* The modern pygmies of Flores Island (November 6, 2018).
* What if yeast had only one chromosome? (August 26, 2018).
* Ancient DNA and the archeologists (April 27, 2018).
* What do microbes eat when there is nothing to eat in Antarctica? (April 2, 2018).
* A blood test that detects multiple types of cancer (March 30, 2018).
* Genetic clues: Why some monkey species don't get "AIDS" upon infection with the immunodeficiency virus (March 26, 2018).
* Ear lobe genetics: more complicated than you thought (March 23, 2018).
* The biological basis of sanguivory (March 2, 2018).
* Identifying individuals from their genomes: a controversy (December 5, 2017).
* A recent genetic change that enhanced the neurotoxicity of the Zika virus (December 1, 2017).
* Nanopore sequencing of DNA: How is it doing? (November 13, 2017).
* DNA sequencing: the future? (November 7, 2017).
* How much of the human genome is functional? (September 1, 2017).
* Is there useful ancient DNA in the dirt? (August 8, 2017).
* Bats and the coronavirus reservoirs (July 25, 2017).
* Cataloging gene knockouts in humans (July 10, 2017).
* More giant viruses, and some evidence about their origin (June 13, 2017).
* DNA evidence in restaurants: is the fish properly labeled? (June 5, 2017).
* Accumulation of deleterious mutations in the last mammoths (May 6, 2017).
* What can we learn from a five thousand year old corn cob? (March 21, 2017).
* Pangenomes and reference genomes: insight into the nature of species (February 7, 2017).
* The Asgard superphylum: More progress toward understanding the origin of the eukaryotic cell (February 6, 2017).
* Malaria history (January 18, 2017).
* The history of brewing yeasts (October 28, 2016).
* How many species of wolf are there, and why does it matter? (October 16, 2016).
* The lost Neandertal Y chromosome (August 13, 2016).
* What is the minimal set of genes needed to make a bacterial cell? (July 9, 2016).
* Contributions of Neandertals and Denisovans to the genomes of modern humans (July 6, 2016).
* Re-introducing captive animals into the wild: an orang-utan mix-up (June 27, 2016).
* Genetics of the giraffe neck (May 27, 2016).
* Better nanopore DNA sequencing by using better nucleotides (May 6, 2016).
* The genetics of being a "morning person"? (April 15, 2016).
* An armadillo the size of a beetle (April 8, 2016).
* Cheese-making and horizontal gene transfer in domesticated fungi (January 19, 2016).
* The autism-Angelman connection: a single enzyme involved in two brain disorders (November 9, 2015).
* Is the warnowiid ocelloid really an eye? (October 12, 2015).
* On genome duplications (September 10, 2015).
* In the shadow of Ebola: The story of Lassa virus (August 26, 2015).
* Ancient DNA: an overview (August 22, 2015).
* A DNA test that can distinguish identical twins (July 17, 2015).
* Finding a gene behind the beak diversity of Darwin's finches (July 14, 2015).
* Our Loki ancestor? A possible missing link between prokaryotic and eukaryotic cells? (July 6, 2015).
* A person who might, just possibly, have met his Neandertal ancestor (June 30, 2015).
* Who's been genetically engineering the sweet potatoes? (June 28, 2015).
* Humans resistant to arsenic? (June 16, 2015).
* Comparing woolly mammoth genomes over time (June 1, 2015).
* Xystocheir bistipita is really a Motyxia: significance for understanding bioluminescence (May 9, 2015).
* Hybrid formation between organisms that diverged 60 million years ago (May 8, 2015).
* A DNA sequencing machine you can carry with you (April 14, 2015).
* Genes that make us human: genes that affect what we eat (February 18, 2015).
* What can we learn from reading (the DNA from) old parchments? (January 30, 2015).
* How did tuberculosis get to the Americas? (January 24, 2015).
* Can we make sense of the many genes involved in autism? (January 16, 2015).
* Mutations that lead to reduced risk for heart disease (November 21, 2014).
* A 115-year-old person: What do we learn from her blood? (November 18, 2014).
* Ebola virus: ancient origins? (November 4, 2014).
* The emerging role for genome sequencing of sick babies (October 27, 2014).
* Cryptozoology meets DNA: No evidence for Bigfoot or Yeti or such (September 13, 2014).
* The new Ebola virus (August 19, 2014).
* A novel nervous system? (July 20, 2014).
* Airport food: What do the birds eat? (May 24, 2014).
* Chromosomes -- 180 million years old? (April 18, 2014).
* The First Americans: the European connection (February 8, 2014).
* The $1000 genome: we are there (maybe) (January 27, 2014).
* DNA from a 400,000-year-old "human" (December 9, 2013).
* Are there genetic issues that we don't want to know about? (October 22, 2013).
* The rotifers that lack sexual reproduction: what do we learn from the genome? (October 4, 2013).
* More genome sequencing for newborns? (September 17, 2013).
* On a similarity of bats and dolphins (September 15, 2013).
* The largest known virus (August 5, 2013).
* The oldest DNA: the genome sequence from a 700,000-year-old horse (August 4, 2013).
* Are DNA sequencing devices resistant to radiation? And why might we care? (July 16, 2013).
* The spruce genome: it's big (July 1, 2013).
* Tracking the pathogen of the Irish potato blight (June 25, 2013).
* Junk DNA: message from the bladderwort (June 4, 2013).
* Microbes on your fresh fruits and vegetables? (May 29, 2013).
* Sharing microbes within the family: kids and dogs (May 14, 2013).
* It's a dog-eat-starch world (April 23, 2013).
* Bacteria on human teeth -- through the ages (March 24, 2013).
* Using DNA for data storage (March 5, 2013).
* Accumulation of mutations in the sperm of older fathers (November 19, 2012).
* How long does DNA survive? (October 23, 2012).
* Male DNA found in human female brains (October 8, 2012).
* Genome sequencing of a human fetus (August 25, 2012).
* The genome of Musa acuminata (August 8, 2012).
* In humans, rare mutations are common (July 24, 2012).
* Nanopores -- another revolution in DNA sequencing? (June 22, 2012).
* DNA sequencing: an overview of the new technologies (June 22, 2012).
* Tracking illegal fish (June 15, 2012).
* A polar bear update (June 3, 2012).
* The Iceman's blood (May 14, 2012). Added November 16, 2023... An update, at the end, focuses on the genome.
* Leopard horses (December 2, 2011).
* Is whole genome sequencing a useful medical tool? (November 9, 2011).
* DNA testing -- for rejection of a transplanted heart (May 24, 2011).
* Studying cancer development by analyzing the genomes of individual cancer cells (May 16, 2011).
* The $1000 genome: Are we there yet? (March 14, 2011).
* Who is #1: the most DNA? (March 7, 2011).
* The Siberian finger: a new human species? -- A follow-up in the story of Denisovan man (January 14, 2011).
* How many human genomes have been sequenced? (November 30, 2010).
* Plants need bacteria, too (October 9, 2010).
* Genome from Mars (September 22, 2010).
* The Siberian finger: a new human species? (April 27, 2010).
* The human genome - 10 years on (April 16, 2010).
* Genome sequencing to diagnose child with mystery syndrome (April 5, 2010). Links to an update.
* Inuk, a 4000 year old Saqqaq from Qeqertasussuk (March 1, 2010).
* The panda genome (January 11, 2010).
* Did Neandertal children hate broccoli? (November 22, 2009).
* The mouse genome (June 5, 2009). This post also mentions the poplar genome.
* A mammoth story (December 1, 2008).
top of page
Miscellaneous articles (2007-2010)
This section used to be "Recent items, briefly noted".
There is more about genome issues on my page of Molecular Biology Internet Resources. In particular, there is a section on Genomes; it includes information on sequencing technologies, as well as collections of genome information.
Also see previous section. That section largely superseded this one, though there is some date overlap.
Topics listed are in order by date, most recent first. If there are multiple items within a topic, the topic is listed by the most recent item.
Coumadin (warfarin) is a widely prescribed medication to reduce blood clotting. The dosage must be carefully controlled, and people vary in how they respond. The FDA has announced a new labeling of coumadin that encourages testing the patient for two known genetic factors that affect the metabolism of the drug. A brief version of the announcement is at https://web.archive.org/web/20170118091648/http://www.fda.gov/Safety/MedWatch/SafetyInformation/SafetyAlertsforHumanMedicalProducts/ucm152972.htm. (August 16, 2007; now archived.)
A small trial has been reported showing that such testing is beneficial.
* News story: Gene test cuts complications from blood thinner warfarin (March 16, 2010). https://usatoday30.usatoday.com/news/health/2010-03-16-warfarin-gene_N.htm.
* Paper: Warfarin Genotyping Reduces Hospitalization Rates -- Results From the MM-WES (Medco-Mayo Warfarin Effectiveness Study). R S Epstein et al, Journal of the American College of Cardiology 55:2804, June 22, 2010. https://www.onlinejacc.org/content/55/25/2804.
Sequencing technology -- and cost. The human genome project cost about $3 billion. Much technology was developed along the way; as the project wrapped up, it was estimated that one could sequence a person's genome for a few million dollars. There is a dream -- and goal -- of sequencing an individual's genome for a thousand dollars. That may still be a way off, but the cost of sequencing has been declining, in large part due to fundamentally new approaches to sequencing. 2009 brings a report of a complete human genome for $50,000. A news story on this: Cost of Decoding a Genome Is Lowered. A Stanford engineer has invented a new technology for decoding DNA and used it to decode his own genome for less than $50,000. August 10, 2009. https://www.nytimes.com/2009/08/11/science/11gene.html.
Using genetic information to assess risk and guide screening. Most genes that affect disease susceptibility have only a small effect. How do we use such information? A paper in the New England Journal of Medicine lays out a model. Although there is probably much to quibble with, the model is clear enough, and may be a useful reference point for discussion. They start with the current UK recommendation that women be screened for breast cancer starting at age 50. Accepting this as the starting point, they note that this is the point at which a woman has a 2.3% chance of breast cancer within the next 10 years. They then argue that by a simple test for some known genetic variants, they can mark some women for screening at age 40 -- because with their genetic makeup that is the age at which they now have a 2.3% risk of breast cancer within 10 years. Similarly, women with other genetic variants have lower risk, and their screening can be delayed. The result is the same use of resources, but more effectively deployed. A news story about this work: Cancer gene test 'for all women', June 26, 2008. Online: http://news.bbc.co.uk/1/hi/health/7475312.stm. The paper is P D P Pharoah et al, Polygenes, Risk Prediction, and Targeted Prevention of Breast Cancer. N Engl J Med 358:2796, 6/26/08. Free online: https://www.nejm.org/doi/full/10.1056/NEJMsa0708739.
Genome ethics. Genome work is raising a new set of ethics questions -- especially since there is so much uncertainty what the genome information means at this point. A group of bioethicists has proposed a set of guidelines for doing genome research, published as: T Caulfield et al, Research Ethics Recommendations for Whole-Genome Research: Consensus Statement. PLOS Biology 6, e73, 3/08. The paper is free online: https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0060073.
Ancestry. An interesting subject is tracing human lineages by genetic tests. This is indeed a proper area of study, and has yielded insights into human migrations. It has also entered the popular arena. There are commercial tests that claim to reveal your ancestry. Unfortunately, the quality of this testing is questionable at this point. A "Policy Forum" article about this appeared in Science, and a news story about the work and that article appeared in the UC Berkeley news. The Science article: D A Bolnick et al, Genetics: The science and business of genetic ancestry testing. Science 318:399, 10/19/07. The UC Berkeley news story, featuring co-author Kimberly TallBear: Researchers caution against genetic ancestry testing; October 18, 2007. https://www.berkeley.edu/news/media/releases/2007/10/18_genetictesting.shtml.
Craig Venter is one of the pioneers of genome work. He is also the first person to have his entire DNA -- the diploid chromosome set -- completely sequenced and reported. Importance? Well, for now it is a technical milestone and something of a curiosity. However, as more complete genomes become available -- and as the cost comes down -- the usefulness will increase. For example, they note how he has specific alleles that both favor and disfavor heart disease. At this point, that is too little info to be useful. At some point, with more information, it will be useful. I doubt that many will want to read this in detail, but simply browsing the Introduction and Discussion sections will give the flavor. And it is a historic paper. The paper -- by Venter, about Venter, and from the Venter Institute -- is: S Levy et al, The Diploid Genome Sequence of an Individual Human. PLoS Biol 5(10):e254. 9/4/07. It is open access at https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050254.
Top of page
BITN home page (Includes list of all BITN topics.)
List of pages of Internet resources
Contact information
Site home page
Last update: November 15, 2024

