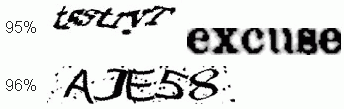Home
> Musings: Main
> Archive
> Archive for September-December 2013 (this page)
| Introduction
| e-mail announcements
| Contact
Musings: September-December 2013 (archive)
Musings is an informal newsletter mainly highlighting recent science. It is intended as both fun and instructive. Items are posted a few times each week. See the Introduction, listed below, for more information.
If you got here from a search engine... Do a simple text search of this page to find your topic. Searches for a single word (or root) are most likely to work.
Introduction (separate page).
This page:
2013 (September-December)
December 30
December 17
December 11
December 4
November 26
November 20
November 13
November 6
October 30
October 23
October 16
October 9
October 2
September 25
September 18
September 11
Also see the complete listing of Musings pages, immediately below.
All pages:
Most recent posts
2026
2025
2024
2023:
January-April
May-December
2022:
January-April
May-August
September-December
2021:
January-April
May-August
September-December
2020:
January-April
May-August
September-December
2019:
January-April
May-August
September-December
2018:
January-April
May-August
September-December
2017:
January-April
May-August
September-December
2016:
January-April
May-August
September-December
2015:
January-April
May-August
September-December
2014:
January-April
May-August
September-December
2013:
January-April
May-August
September-December: this page, see detail above
2012:
January-April
May-August
September-December
2011:
January-April
May-August
September-December
2010:
January-June
July-December
2009
2008
Links to external sites will open in a new window.
Archive items may be edited, to condense them a bit or to update links. Some links may require a subscription for full access, but I try to provide at least one useful open source for most items.
Please let me know of any broken links you find -- on my Musings pages or any of my regular web pages. Personal reports are often the first way I find out about such a problem.
December 30, 2013
3D printing: Sculplexity -- and a printed model of a forest fire
December 29, 2013

|
A forest fire. Or, rather, a model of a forest fire. A 3-dimensional model.
The model is about 8 centimeters (3 inches) on a side.
This is trimmed from the top figure in the Kurzweil news story. The same figure appears widely. It is related to Figure 10 of the article.
|
What's this about? It's another example of a clever application of 3D printing. The lead scientist behind this development, a theoretical physicist, suggests that such printed models can serve to illustrate complex ideas of theoretical physics. The forest fire, reported in a new paper and shown above, is just an example.
I'm not sure I find the model of a forest fire, pictured above, all that useful. But that misses the point. The article introduces a tool: printed 3D models to illustrate complex ideas. We'll see what people make of this new tool.
The article also introduces, in its title, a new word.
News stories:
* Using 3D printing to explain theoretical physics. (Kurzweil, December 10, 2013.)
* 3D printing used as a tool to explain complex theoretical physics. (3Ders, December 9, 2013.)
* Sculplexity: sculptures of complexity using 3D printing. (Centre for Complexity Science (Imperial College London), December 5, 2013. Now archived.) A blog post, by one of the authors of the article (Evans).
The article: Sculplexity: Sculptures of Complexity using 3D printing. (D S Reiss et al, European Physics Letters, 104:48001, December 9, 2013.) Check Google Scholar for a freely available copy.
The article and some of the news stories also contain a photograph of a coffee table -- a work of art that emerged from a 3D printer and served to stimulate the current development. It's quite striking.
More about 3D printing:
* 3D printing: Make yourself a model of the universe (December 19, 2016).
* 3D printing: simple inexpensive prosthetic arms (January 29, 2014).
* 3D printing: Neurosurgeons can practice on a printed model of a specific patient's head (December 16, 2013).
More about 3D vision: What can we learn by giving a praying mantis 3D glasses while it watches a movie? (March 12, 2016).
A "flower" that bites -- and eats -- its pollinator
December 27, 2013

|
Do you like orchids?
What do you think? Would you be tempted to pollinate this "flower"?
Many an insect has tried -- only to be eaten, as this mantis reveals itself. What seems to be a flower, perhaps an orchid, is actually an insect. A mantis, one known as the orchid mantis.
This is trimmed and reduced from the first figure in the National Geographic news story. The width of the figure probably represents about 5-10 centimeters.
|
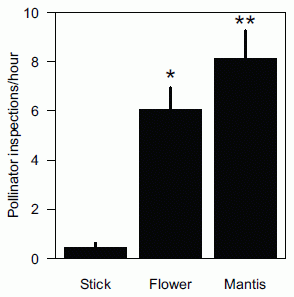
How well does this deception work? Here is a test, as reported in a new article...
In this experiment, scientists attached three candidate "flowers" to natural outdoors vegetation. They observed the frequency of visits of pollinating insects. The graph shows that frequency of pollinator visits for the three "flowers".
You can see that the pollinators made few visits to the stick, but made many visits to both the real flower and the orchid mantis. In fact, in this test, the results were highest with the mantis. The details might depend on the specific flowers used; the point is that the mantis was an attractive target for the pollinating insects.
This is Figure 3 from the article.
|
|
The idea that the orchid mantis poses as a flower to attract its food is not new. What's new is that scientists have now provided some good evidence -- including video footage -- for this behavior. Their little movie, which accompanies the article, is listed below.
News stories:
* Study shows orchid mantis more attractive to their prey than real orchids. (Phys.org, October 15, 2013.)
* Praying Mantis Mimics Flower to Trick Prey. (National Geographic, September 25, 2013. Now archived.)
Movie (1 minute; no sound).You may need to play it more than once to follow what is going on. (It's also available from the article web site, under "Supplements".)
The article, which is freely available: Pollinator Deception in the Orchid Mantis. (J C O'Hanlon et al, American Naturalist 183:246, January 2014).
More about pollination...
* What if there weren't enough bees to pollinate the crops? (March 27, 2017).
* Caffeine boosts memory -- in bees (April 12, 2013).
Other posts about deception and mimicry include:
* A plant that mimics the leaves of its host (May 30, 2014).
* A deceptive robot (September 4, 2012).
* Deceiving a rival male (August 28, 2012).
This post may remind you of carnivorous plants, such as in the post Carnivorous plants: A blue glow (March 16, 2013). But remember, the orchid mantis is an animal that mimics a plant; it is not a plant.
More about mantises:
* Can an insect catch fish for its dinner? (October 8, 2018).
* What can we learn by giving a praying mantis 3D glasses while it watches a movie? (March 12, 2016).
More about flowers... pH and the color of petunias (March 26, 2014).
Also see... How the tomato plant resists the Cuscuta (November 4, 2016).
December 17, 2013
3D printing: Neurosurgeons can practice on a printed model of a specific patient's head
December 16, 2013
Imagine being able to practice neurosurgery. More specifically, imagine being able to practice on a particular patient's brain tumor.
A new article reports significant progress towards that goal. The general idea is to take a CT scan of the patient's head, then print a model of the head, using the latest in 3D printers.
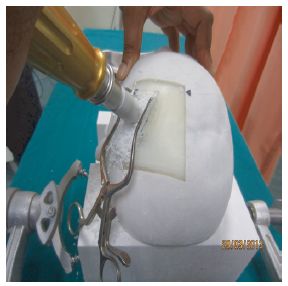
|
Practicing brain surgery.
A hole is drilled through the skull -- of a model of the patient's head, made by 3D printing.
This is Figure 3 from the article.
|
The key advances in the new work were in improving the ability to print multiple materials, with different properties. As a result, the printed model head has more realistic bones and tissues.
These models can be used during training of neurosurgeons. Further, models of a specific patient's brain, including its tumor or other pathology, can be used for practicing the treatment of a particular patient.
How much do these models cost? The key part, which must be replaced for each surgery, is about $600. The authors think that is in the range where such models will be practical for training. Of course, further cost decreases are likely.
Movie. There is a 2 minute movie illustrating the model. It is linked within the article. You can see the movie directly at: file for Windows Media Player or file for Quicktime.
News story: Multi-material 3D printer creates realistic neurosurgical models for training. (Kurzweil, December 12, 2013.) Includes a picture of the printer.
The article: Utility of multimaterial 3D printers in creating models with pathological entities to enhance the training experience of neurosurgeons. (V Waran et al, Journal of Neurosurgery 120:489, February 2014.)
More about brain surgery: Coupling the surgeon's knife to a mass spectrometer (August 13, 2013).
More about 3D printing:
* 3D printing: Make yourself a model of the universe (December 19, 2016).
* 3D printing of human tissues: the ITOP (May 24, 2016).
* 3D printing: simple inexpensive prosthetic arms (January 29, 2014).
* 3D printing: Sculplexity -- and a printed model of a forest fire (December 29, 2013).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Brain/A>. It includes a list of brain-related Musings posts.
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Cancer. It includes a list of some other Musings posts on cancer.
Chelation therapy -- a controversial clinical trial
December 13, 2013
A chelator is a substance that binds metal ions, such as Ca2+. Chelators are used in medicine to treat some types of metal poisoning, such as lead (Pb) poisoning.
The possible use of chelators to treat other conditions, such as heart disease, has been suggested. There is neither good data to support such a use, nor any understanding of why it might work. However, chelation therapy to treat heart disease is used by some, as what we sometimes call alternative medicine.
The US National Institutes of Health (NIH) has devoted some effort to studying alternative medicine treatments, with the intent of providing good data about such treatments, pro or con. An example is the Trial to Assess Chelation Therapy (TACT). This major clinical trial was designed to test whether a particular chelation therapy might be a useful treatment for heart disease. The trial itself has become the subject of controversy. In a short space here, I can only begin to describe the story. I emphasize at the start, then, that the goal is to present some of the issues, not to reach a conclusion.
The basic plan of the trial was to treat patients with a chelator or a placebo, and measure the frequency of heart-related adverse outcomes. The trial was reported in March 2013. The basic finding was a small improvement associated with chelation therapy. The improvement was not statistically significant, but perhaps deserved further study. The trial itself was criticized for various problems, which skeptics thought biased the trial toward showing a positive result for chelation.
The authors of the original article on the trial now have a new article, with further analysis of the data. In particular, they report that if they consider only those patients who had diabetes, there was a large beneficial effect of chelation therapy.
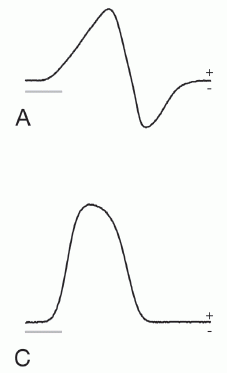
Here are those results...
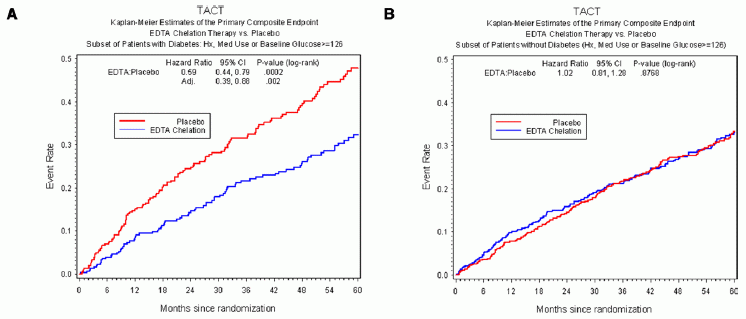
Each frame shows adverse events (y-axis) vs time (x-axis), for those treated with placebo (red curve) or the EDTA chelation treatment (blue curve). Frame A (left side) is for patients with diabetes; frame B (right side) is for patients without diabetes.
The "event rate" shown on the y-axis is apparently the cumulative number of adverse events, expressed as a decimal fraction of the number of patients in the group. The x-axis scale shows time in "months since randomization"; it is time since the start of the treatment. The treatment lasted about one year. (Some of the details are not clear in this article, but are presumably in the earlier article.)
The main observation is clear. For patients without diabetes (frame B, right side), there is no difference between placebo and chelation. For patients with diabetes (frame A, left side), there are many fewer events in the group with chelation therapy. The difference is statistically significant.
The event rate curve indicates that the benefit extended beyond the treatment time. In fact, the curve is substantially linear.
A bit of fine print...
The target population for the trial was people who had already suffered one heart attack.
The chelator used here is a well-known chemical commonly known as EDTA. The full name is ethylenediamine tetraacetic acid. It is widely used in chem labs as a chelator.
This is Figure 2 from the article. (Readability of the text in the figure was not very good in the original.)
|
What do we make of this? The new article makes a case that the chelation therapy is of benefit to those with diabetes. However, it is generally considered improper to reach conclusions for tests not planned as part of the original trial. The problem is that, once you have the data, you can look around. If you look at enough things, you will find some that test as significant. The idea of statistical probability requires that. Thus the proper conclusion is to take the diabetes result as a "clue" -- one that should be tested further. The authors handle this well; they are quite explicit that one cannot consider the new analysis "proof" of effectiveness for those with diabetes. However, it is human nature that some will over-interpret these results. Further, claims of problems with the trial remain.
Thus the situation is not very satisfying. After all this effort, we still don't know if chelation is an appropriate therapy for heart disease, perhaps in those with diabetes.(It does now seem unlikely that it is of benefit for those without diabetes.) In one sense, this is all normal science. Results in science are often inconclusive, and need follow-up. But this is medicine; people's lives are at stake. Further, this was a clinical trial that was designed to resolve a question. It didn't.
News story: Chelation Therapy Reduces Cardiovascular Events for Older Patients with Diabetes. (NIH, November 19, 2013. Now archived.) From the funding agency and sponsor.
The article, which is freely available: The Effect of an EDTA-based Chelation Regimen on Patients With Diabetes Mellitus and Prior Myocardial Infarction in the Trial to Assess Chelation Therapy (TACT). (E Escolar et al, Circulation: Cardiovascular Quality and Outcomes 7:15, January 2014.)
As noted, the current paper is a follow-up to the original report on this clinical trial. That was published in JAMA in March. It is reference 5 of the current article, and is freely available at the JAMA site. Of particular interest here is that the original article was accompanied by two editorials. One was from the JAMA editor, discussing (and defending) the decision to publish; the other was from a skeptic. It is interesting to look over both of them. Do remember that these are on the original report, not the current article. Each is two pages.
Editorials on original article; both are freely available:
* Evaluation of the Trial to Assess Chelation Therapy (TACT) -- The Scientific Process, Peer Review, and Editorial Scrutiny. (H Bauchner et al, JAMA 309:1291, March 27, 2013.)
* Concerns About Reliability in the Trial to Assess Chelation Therapy (TACT). (S E Nissen, JAMA 309:1293, March 27, 2013.)
This is not our first story of concerns about clinical trials. Also see...
* The missing clinical trials data: it's time to RIAT (July 22, 2013).
* Golden rice as a source of vitamin A: a clinical trial and a controversy (November 2, 2012).
See Mission Improbable (November 10, 2009) for more about the statistics of clinical trials.
More about heart disease... Red meat and heart disease: carnitine, your gut bacteria, and TMAO (May 21, 2013).
For more about diabetes, see my Biotechnology in the News (BITN) topic Diabetes. It includes a listing of some other Musings posts in the area.
More about placebos... Would a placebo work even if you knew? (January 31, 2014).
More about chelation: The magnesium dilemma: a step toward understanding how RNA might have been made in "protocells" (February 22, 2014).
December 11, 2013
Could vibration (or loud music) improve the performance of a solar cell?
December 11, 2013
A solar cell uses light energy to excite electrons. The excited electrons are then used in some way; for example, they may become part of an electric current. Is it possible that vibrating a solar cell would improve its performance? A new article suggests it might, and offers an explanation for why. The article also contains an amusing aspect, which has gotten news media attention.
In this work, the scientists use solar cells that include nanorods made of zinc oxide, ZnO. An interesting feature of ZnO is that it is piezoelectric: vibration induces electrical current in ZnO.
In one simple experiment, they measured the efficiency of the solar cell with ZnO nanorods as they also provided acoustical vibration (sound; loosely, noise). They tested sound over a wide frequency range, up to 50 kilohertz (kHz).
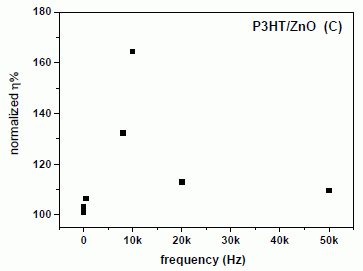
|
The figure at the left shows the results. "P3HT" is the polymer coating that is the actual light receptor.
The graph plots the efficiency of the solar cell (y-axis) vs the sound frequency used (x-axis). The efficiency is on a relative scale; 100% is the baseline efficiency without added vibration. (See the point with no sound, at 0 Hz; it is 100%.)
You can see that providing vibration at about 10 kHz improves the efficiency of the solar cell by about 50%.
This is Figure S4 from the Supporting Information with the article.
|
What's going on here? One possibility is that the ZnO piezoelectricity induced by the noise is simply being added onto the solar-induced current. The numbers do not support this; the enhancement is more than can be accounted for by the piezoelectricity.
The scientists do further tests, and suggest a more interesting mechanism, one that gets to the heart of how solar cells work -- and why they don't work very well. As noted, the solar energy is used to excite an electron. Where the electron used to be (where it gets excited from, we might say) is called a "hole". That is, the solar excitation creates an electron-hole pair. The secret of solar cell efficiency is to get that electron out into an electrical circuit. If the electron simply drops back into its hole (or any other hole it might encounter), nothing is gained. It seems, according to their analysis, that excited electrons are less likely to drop back into holes in the presence of the acoustic stimulation. That is, the effective lifetime of excited electrons is increased, making it more likely that they will be captured as useful energy. How does the noise reduce electron-hole recombination? That they don't know, but they suggest it has to do with the electric field set up by the piezoelectricity of the ZnO.
Is this useful? I think it's too early to know. The solar cells they are using are quite inefficient, far below current standards, even with vibration. Would the vibration effect be useful with any solar cell of good efficiency? That's open. Regardless, it would seem that the effect may provide some insight into the issue of electron-hole recombination.
I noted that there was an amusing aspect to the work, one noted by the news media. Here is a paragraph from the article, very near the end (next to last paragraph of the main body of the paper, just before the "Experimental Section")... "On a lighter note, the response of the devices to a variety of acoustic conditions was investigated by playing a variety of different types of music during testing, rather than single frequency signals as used above. It was found that the efficiency enhancement was most pronounced for pop rather than classical music, most probably due to the increased amplitudes of higher frequencies typically present in electronically synthesised music."
News stories:
* Playing Pop and Rock Music Boosts Performance of Solar Cells. (Science Daily, November 6, 2013.)
* Jingle cells are rocking on sunshine. (Chemistry World, Royal Society of Chemistry, November 6, 2013.)
The article, which is freely available: Acoustic Enhancement of Polymer/ZnO Nanorod Photovoltaic Device Performance. (S Shoaee et al, Advanced Materials 26:263, January 15, 2014.)
Other posts about solar energy include...
* Solar cells: a new record for efficiency (May 26, 2020).
* Solar energy: A more efficient way to boil water? (September 12, 2014).
* An artificial forest with artificial trees (June 7, 2013).
* Making electricity in your windows: sharing the solar spectrum (July 5, 2011).
More about ZnO nanorods... Electronic devices that can work under water (November 7, 2011).
There is more about energy on my page Internet Resources for Organic and Biochemistry under Energy resources. It includes a list of some related Musings posts.
There is more about music on my page Internet resources: Miscellaneous in the section Art & Music. It includes a listing of music-related Musings posts.
DNA from a 400,000-year-old "human"
December 9, 2013
Stories of ancient DNA continue to amaze -- and to confound.
We now have a report of a DNA sequence from a hominin -- a human-like animal, in the broad sense -- that lived about 400,000 years ago. It's just the mitochondrial DNA, but still it is quite remarkable. It's several times older than any previous hominin DNA sequence.
What do we learn from this new hominin's DNA? That's not at all clear. We'll note the key result in a moment, but I think this is a good time to remind readers that science is often tentative. The new article is, in some ways, a breakthrough, but the results are thin at this point. It is best to take the new results as preliminary.
The following figure is a map, showing the sources of ancient DNA samples prior to this new work. Red dots (sites 3-14) are for Neandertal samples and a single blue dot (site 2, at the right) is for Denisovan DNA.
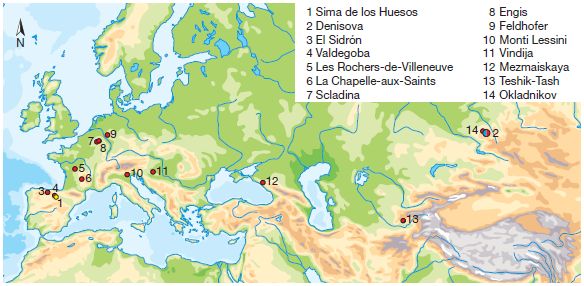
|
Site 1 (lower left, yellow dot) is the source for the new work. It's "Sima de los Huesos" ("pit of bones"), in northern Spain.
This is Figure 1 from the article.
|
And the new DNA? It seems most closely related to Denisovan DNA.
That's not what was expected -- as you might guess by looking at the map.
You'll get a sense of the range of reactions of scientists if you look over the news stories. But let's relax. This is one sample. It's the first sample of seemingly Denisovan DNA outside Denisova. Who knows how typical this sample is, or even if the data are entirely correct. And who knows what the distribution of Denisovans was, given that we had only one previous example.
This is an exciting piece of work. It is, after all, the oldest genome sequence on the ancient hominin lineages. However, it is too early to conclude much. Let's just sit back and wait for more data. Indeed, the news stories -- and the final paragraph of the article -- hint that more analyses from this site, including nuclear DNA, may be forthcoming.
News stories:
* Oldest Hominin DNA Sequenced: Mitochondrial Genome of a 400,000-Year-Old Hominin from Spain Decoded. (Science Daily, December 4, 2013.)
* Hominin DNA baffles experts -- Analysis of oldest sequence from a human ancestor suggests link to mystery population. (Nature News, December 4, 2013.)
The article: A mitochondrial genome sequence of a hominin from Sima de los Huesos. (M Meyer et al, Nature 505:403, January 16, 2014.) Put the title in Google Scholar; you may find a freely available pdf of the article.
For more about Denisovan DNA: What is Denisovan man telling us about Asian-Pacific man? (October 25, 2011).
This is not the oldest genome yet sequenced. That is discussed in the recent post The oldest DNA: the genome sequence from a 700,000-year-old horse (August 4, 2013).
More about old human genomes: The First Americans: the European connection (February 8, 2014).
There is more about genomes on my page Biotechnology in the News (BITN) - DNA and the genome. It includes an extensive list of Musings posts on sequencing and genomes.
A book on ancient human genomes... See my page of Book suggestions: Pääbo, Neanderthal Man -- In search of lost genomes (2014).
More about Spain: Flow centrality: the key to a scientific analysis of the soccer game (July 11, 2010).
Was Linnaeus's original elephant African or Asian?
December 7, 2013
Carl Linnaeus was the 18th century biologist who introduced the modern approach to systematic classification and naming of organisms. The common genus-species "binomial" name, such as Homo sapiens for modern humans, comes from the Linnaeus system. Or Elephas maximus...

|
There it is, in a recent picture. It is the elephant that Linnaeus designated, in 1758, as the type specimen for elephants. The "type specimen" is the specific example that has been chosen to represent the type of organism.
The scale bar is "approximately 10 cm". It's a small elephant. In fact, it is a fetus. The code at the bottom is the specimen number from the Swedish Museum of Natural History.
This is Figure 1B from the article.
|
You probably know that there are two kinds of elephants, quite distinct: African elephants and Asian elephants. So, which was it? Was Linnaeus's original elephant African or Asian?
That turns out to be an interesting story. Linnaeus himself did not make the distinction. He simply recognized the "elephant", and this was his type specimen. Only later was the distinction between African and Asian elephants made. The elephant shown above was considered to be an Asian elephant. Asian elephants got the name Linnaeus had given -- and they got the type specimen.
Many have questioned that assignment over the years. A new article presents a rather thorough analysis, proving "beyond a doubt" that this was an African elephant. This is based on multiple morphological features, as well as DNA and protein sequences. The article then reassigns the original type specimen, long considered an Asian elephant, to be the type specimen for African elephants. It further finds an appropriate specimen to now serve as a proper type specimen for Asian elephants.
News story: Centuries-Old Elephant Imposter Unmasked. (Science Daily, November 5, 2013.) A delightful news story, summarizing both the historical and biological aspects.
The article: Resolution of the type material of the Asian elephant, Elephas maximus Linnaeus, 1758 (Proboscidea, Elephantidae). (E Cappellini et al, Zoological Journal of the Linnean Society 170:222, January 2014.) (Put the title into Google Scholar, and you may find a copy of this article freely available.)
Some of the news coverage of this story has been misleading, in suggesting that Linnaeus mis-classified the specimen. That's not really fair. As noted above, he did not distinguish types of elephants. Others, later, classified the specimen as Asian, which we would now consider incorrect. In any case, re-classification of organisms occurs from time to time, as better information becomes available. There is no blame here; it is a story of progress -- and a fun story at that.
More about elephants:
* Do elephants suffer long term harm if their social groups are disrupted by human intervention? (April 27, 2014).
* Atomic bombs and elephant poaching (October 25, 2013).
* If the elephant can't find its dinner, should you help by pointing to it? (October 18, 2013).
A recent post discussed how humans should be classified into species, and raised the possibility that some current views on human ancestors may be wrong. As with the current post, the issue is sorting through all we know, and deciding which criteria are most important. It's not simple. An interesting skull, and a re-think of ancient human variation (November 12, 2013).
You did notice the name of the journal for the article here? It's appropriate for an item about Linnaeus. Here is a previous post from another journal of the Society... Who is #1: the most DNA? (March 7, 2011).
More Linnaeus: Top 10 new species for 2015 (June 3, 2015).
Progress toward an artificial fly
December 6, 2013
Briefly noted...

|
Artificial flies.
Weight: about 80 milligrams.
This is Figure 1A from the article.
|
Two movie files do a good job of showing what has been accomplished. The movie files are posted at the journal web site with the article; choose Supplementary Materials. The movies are freely available regardless of whether you have subscription access to the article itself. Each movie is about 2 minutes; no sound. Each movie shows a sequence in real time, then repeats it at a slower speed. The main focus of movie S1 is to show the fly hovering; the main purpose of movie S2 is to show lateral maneuvers.
The development of the artificial fly required technological advances in miniaturization, control circuitry, and manufacturing. The news stories highlight some of the issues -- and limitations. (A key limitation is evident at the end of each video clip.) They note that, whether the flies themselves are of use (artificial pollinators?), the advances made while developing them may be important.
News stories:
* A robotic insect makes first controlled test flight. (Kurzweil, May 9, 2013.)
* Robotic insects make first controlled flight. (Wyss Institute, Harvard, May 2, 2013.) From the Institute where the work was done.
The article: Controlled Flight of a Biologically Inspired, Insect-Scale Robot. (K Y Ma et al, Science 340:603, May 3, 2013.) Includes two movie files, as noted above.
What if there weren't enough bees to pollinate the crops? (March 27, 2017). Another step toward developing an artificial pollinator.
Another example of a robot designed with an insect in mind: Acrobatic cockroaches inspire robot design (September 16, 2012).
For more about bio-inspired design, see my Biotechnology in the News (BITN) topic Bio-inspiration (biomimetics). It includes a listing of some other Musings posts in the area.
More about robots with novel locomotion: Cubli: a little cube that can stand on one corner and can walk (January 14, 2014).
More about flies:
* A superhydrophobic fly -- that can survive in highly alkaline water (February 25, 2018).
* "Moonwalkers" -- flies that walk backwards (May 28, 2014).
* Making smarter flies (July 18, 2012).
More about flying:
* How to fly a beetle (April 27, 2015).
* Why don't penguins fly? (August 24, 2013). Both this and the current post deal with the energy costs of flying.
A book about flying is listed on my page Books: Suggestions for general science reading. Alexander, On the Wing -- Insects, pterosaurs, birds, bats and the evolution of animal flight (2015).
December 4, 2013
Characterization of carbon nanotubes
December 3, 2013
Carbon nanotubes (CNTs) are tiny tubes made up of thin sheets of graphite-like carbon. The walls of CNTs are one or a few atoms thick, as with graphene. That is, you can think of CNTs as being like sheets of graphene rolled up into a tube. Scientists have been making CNTs for years, and studying their properties and possible technological applications. A major barrier to use of CNTs is that there are different types, and it is hard to tell them apart or separate them. For example, there are CNTs that conduct electricity and those that do not (or are semi-conductors).
A new article offers an advance in seeing CNTs, and -- more importantly -- in characterizing them.
The scientists use a light microscope -- a complex light microscope. They have developed technology that allows them to use polarized light with high spatial resolution. The spatial resolution is needed to see the tiny tubes; the polarized light allows them to distinguish the types of CNTs.
Here is a carbon nanotube, as observed with their light microscope...

|
It's that horizontal line about half way down.
That may not look like much, but it is an impressive achievement to see a CNT like this. It is only a nanometer or so in diameter.
Analysis of the polarized light in this case shows that this CNT is a semi-conductor.
|
This is Figure 2g from the article. The full figure includes diagrams, optical and EM images, and polarized light analyses -- for CNTs in different environments. (The CNT above is simply lying on a piece of silica, SiO2.) More of that full figure is also in both news stories listed below. (The figure above represents about 25 µm across; scale bars are in other parts of the full figure.)
|
What makes this important is that the method allows them to scan large numbers of CNTs, and determine the properties. This will be useful in research labs. The ultimate goal is to learn how to make CNTs with consistent properties.
News stories:
* First images and spectra of individual carbon nanotubes in a general environment. (Kurzweil, November 14, 2013.)
* From the Lab: Taking a New Look at Carbon Nanotubes -- Berkeley Researchers Develop Technique For Imaging Individual Carbon Nanotubes. (Lawrence Berkeley National Laboratory (LBNL), November 12, 2013.) (The work is largely from the Physics Department at UC Berkeley and LBNL.)
* News story accompanying the article: Carbon nanotubes: Captured on camera. (M W Graham, Nature Nanotechnology 8:894, December 2013.)
* The article: High-throughput optical imaging and spectroscopy of individual carbon nanotubes in devices. (K Liu et al, Nature Nanotechnology 8:917, December 2013.) There is a preprint available at ArXiv: ArXiv preprint.
More about CNTs...
* How do you get silkworms to make stronger silk, reinforced with graphene? (October 24, 2016).
* Supercapacitors in the form of stretchable fibers -- suitable for clothing (May 2, 2014).
* A box that will fold up upon command -- heat- or light-actuated switches (September 3, 2011).
* Stanford scientists discover that ink sticks to paper (May 29, 2010).
* Weighing gold atoms (September 24, 2008).
As noted above, graphene and CNTs are related: both consist of one or a few sheets of graphitic carbon. A previous post on graphene: Loudspeakers: From gold-coated pig intestine to graphene (April 27, 2013).
More about advances in light microscopy: A more powerful method for measuring what is in a cell (July 23, 2013).
... including simple microscopy: A ream of microscopes for $300? (June 22, 2014).
Also see a section of my page Internet resources: Biology - Miscellaneous on Microscopy.
More about polarized light... Stripes protect zebra against horseflies -- another story of polarized light (February 26, 2012).
More about high resolution distance measurements... Can the naked human eye measure distance to nanometer accuracy? (July 20, 2015).
This post is listed on my page Introduction to Organic and Biochemistry -- Internet resources in the section on Aromatic compounds. That section includes a list of posts on graphene and carbon nanotubes.
Should physicists be allowed to use lead from ancient Roman shipwrecks?
December 2, 2013
Why do physicists want to use lead (Pb) from ancient Roman shipwrecks? Because it has lower levels of radioactivity, as a result of being isolated for two millennia following purification of the lead. That matters for the physicists. They use the lead as shielding in ultra-sensitive experiments, such as those looking for dark matter. The lower the background radiation from the lead shielding, the better. If there is validity to the need of the physicists, how do we balance that with preserving cultural artifacts?
It's an interesting question, one that I hadn't thought of. It's addressed in a recent article, which is summarized in the news story. I encourage you to read at least the news, and perhaps browse the article, so you see what some of the issues are. Caution... I don't think enough information is given to reach a clear conclusion. Enjoy the question, but resist trying to resolve it.
News story: Controversy over the use of Roman ingots to investigate dark matter and neutrinos. (Phys.org, November 29, 2013.)
The article is based on a talk; it is described as an "extended abstract". The article is freely available; here is a direct link to the pdf: Experiments on Particle Physics Using Underwater Cultural Heritage: The Dilemma. (Elena Perez-Alvaro, Experiments on Particle Physics Using Underwater Cultural Heritage: The Dilemma, Rosetta 13:40, 2013. The talk is from April 2013; the journal issue is probably Fall 2013.) I suspect that it has not been peer reviewed. The article is short and, mostly, easy to follow. [Here is a link to the journal issue, if you want more background about it: the journal.]
More about shipwrecks...
* A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
* An ancient navigation device? (April 16, 2013).
More about dark matter:
* What if there isn't any dark matter? Is MOND an alternative? (December 12, 2016).
* Where is the dark matter? (May 11, 2012). Includes some brief background on the dark matter problem.
More about neutrinos: IceCube finds 28 neutrinos -- from beyond the solar system (June 8, 2014).
More about lead: Lead-rich stars (August 30, 2013).
More about radioactivity: Berkeley RadWatch: Radiation in the environment (February 24, 2014).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Ethical and social issues.
My page of Introductory Chemistry Internet resources includes a section on Nucleosynthesis; astrochemistry; nuclear energy; radioactivity. That section contains some resources on the effects of radiation.
November 26, 2013
Computer reads CAPTCHAs with 90% accuracy
November 25, 2013
That's better than many people can do.
CAPTCHAs? They are those funny characters you need to decipher before you can submit something on a computer. The acronym stands for Completely Automated Public Turing tests to tell Computers and Humans Apart. The name is clear enough. You submit your interpretation of the CAPTCHA to prove to the computer you are human; computers can't read them.
Make that, couldn't read them. Stanford University scientists have announced a computer program that can consistently read them well.
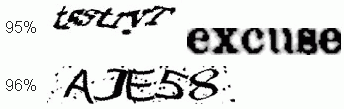
|
Two of the CAPTCHAs that the computer read. Percentage success -- of the computer -- is shown at the left.
This is part of the figure in the news story.
|
This is an interesting development regarding computer submissions. What next? Harder CAPTCHAs? But there is a more serious aspect. The heart of the program is an improved ability of the computer to interpret patterns and pictures. That is an important development.
All we have for now is a news story, so we'll leave it at that. The news story is good, and includes some data, and a short video. It also links to an article. Caution, it is an older article; the work discussed in the news story builds on the article, but goes beyond it.
News story: Vicarious AI breaks CAPTCHA 'Turing test'. (Kurzweil, October 28, 2013.) Note that the team has formed a spin-off company, called Vicarious, to develop this work further.
More about computer pioneer Alan Turing, whose name is invoked above: Alan Turing, computable numbers, and the Turing machine (June 23, 2012).
More about the Turing test: Eugene Goostman and his Turing test (June 17, 2014).
Can memories survive if head is lost?
November 23, 2013
Perhaps, according to a new article.
The general experimental approach used in the new work is logically straightforward...
* Establish a memory.
* Remove head.
* Allow new head to grow.
* Test for the original memory.
This work was done with Planaria, a type of flatworm. Some species of Planaria have a remarkable ability to regenerate; the body can be cut into multiple pieces, and each will regenerate an entire animal -- including head and brain.
The following figure gives the idea, and shows worms at various times during the procedure.
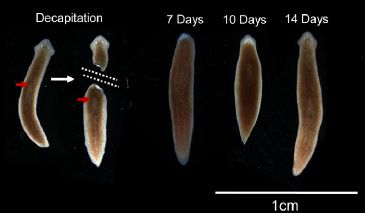
|
An intact worm is cut in two. It is done such that the tail portion lacks all brain structure, so far as is known. The tail portion is allowed to regenerate; by day 14, it has regenerated a rather normal looking complete worm.
This is Figure 4 from the article. It's shown with some of the news stories listed below; the "annotated" version with the Tufts "Total Recall" story is particularly good. (The worms shown are a set photographed together; they are not different views of the same worm at various times.)
|
Those Day 14 worms with regenerated heads were tested. Tested for what? For their ability to do a task they had been trained to do, before losing their head. In fact, a major portion of the work is devoted to developing the experimental system for training and testing planaria.
The figure at the right shows some results. Let's look at the key points.
The y-axis is the time the worms take to do a task. The lower the number, the better.
The x-axis shows there are two types of animals being tested: those that have been trained ("familiarized"; right side), and those that have not been trained ("unfamiliarized" -- the control; left side).
|
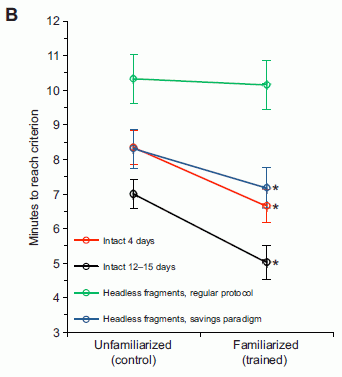
|
Start with the red curve. This is a set of results for animals that were normal, not regenerated. You can see that these "intact" animals did better when they had been trained: the result (the task time) is lower for the familiarized animals. This is a reasonable result. (A second data set with intact animals showed a similar result; black curve.)
Now look at the blue curve. This is for animals that had regenerated a new head. It's rather similar to the red curve, for intact animals. Remember that the training occurred before the original head was removed. That is, the animals being tested had a different head than the one that was trained. This result (blue curve) shows that animals with regenerated heads may retain memories originally acquired with the previous head.
This is Figure 3B from the article.
In discussing the results above, I have skipped one curve -- the green curve. This is also for animals with regenerated heads, and it does not show memory retention. The training procedure for the worms was different in that experiment. That is, it seems that one procedure showed memory retention in the new head, and one did not. I don't understand the procedures very well, so can't comment further. What is important, I think, is that one of two protocols showed memory retention. That's the noteworthy result. It is what they emphasize.
|
How is it possible that a memory might be retained after loss of the brain? At this point, that is entirely speculation. Among the possibilities are that they had not entirely removed the brain (though they took great care to do so), and that memory storage occurs throughout the body.
Convinced? I'm not sure. It's not even clear how sure the authors are. What they emphasize is that they have developed an experimental system to study behavior training and memory in these fascinating animals. That includes the possibility of studying how their behavioral learning is transmitted following the regeneration of missing parts, including the brain. At least some of the results suggest that such memory transmission to a new brain may occur. That's what will be explored in further work.
News stories:
* Decapitated Worms Regrow Heads, Keep Old Memories. (National Geographic, July 16, 2013. Now archived.)
* Researchers discover flat worms retain memories even after decapitation. (Phys.org, July 12, 2013.)
Two stories from Tufts university, where the work was done:
* Flatworms Lose Their Heads but Not Their Memories. (July 18, 2013.)
* Total Recall -- Their heads cut off, worms grow back brains - and retain their earlier memories. (September 25, 2013.)
The article: An automated training paradigm reveals long-term memory in planarians and its persistence through head regeneration. (T Shomrat & M Levin, Journal of Experimental Biology 216:3799, October 15, 2013.)
An example of studying the nervous system of a simple animal: With 24 eyes, can they see the trees? (June 11, 2011).
Another -- simpler, not even an "animal": How slime molds learn (June 11, 2019).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain. It includes a list of brain-related Musings posts.
More about regeneration is on my page of Biotechnology in the News (BITN) for Cloning and stem cells. It includes an extensive list of related Musings posts.
Another flatworm... Could a tapeworm with cancer transmit the cancer to its human host? (November 16, 2015).
November 20, 2013
What has six tails -- and is beyond Mars?
November 20, 2013
There it is, at the right. Its name is P/2013 P5. (You can call it P5, for short.)
The figure is a composite of photos taken by the Hubble Space Telescope (HST), September 10, 2013.
P5 had been discovered, by a smaller telescope, only three weeks earlier.
This is trimmed from a figure from NASA, which has been widely distributed. The full figure shows two such photographs, taken two weeks apart. A version of the figure is in the article, as Figure 1. There the tails are labeled. The color is false.
|

|
What is it? Browse the reference listings below... The article title says it is a comet. The news story from Nature says it is an asteroid. Space Daily titles their news story with a riddle -- and then goes on to suggest it looks like a "lawn sprinkler or badminton shuttlecock".
Based on the evidence so far, we can eliminate the lawn sprinkler. The scientists reporting this are rather sure there is no water.
If you're inclined to accept what the authors say... Read the introduction, in their article. In the first paragraph, they say "The combination of asteroid-like orbit and comet-like appearance together reveal P5 as an active asteroid (equivalently, a main-belt comet -- MBC)."
So, what is the difference between a comet and an asteroid? The old view, simplified, is that comets have highly elliptical orbits that bring them relatively close to the sun, and they have tails that become visible as the sun warms the water-ice of their body. In contrast, asteroids are in orbits between Mars and Jupiter (the asteroid belt), and have rocky bodies. As so often, as we get more observations we find that old classifications break down. In particular, there are objects in the asteroid belt with tails -- quite likely tails of dust rather than ice.
P5 is one of those objects. It's in the asteroid belt, and it has a tail. Six tails, in fact. It's one of the weirder things astronomers have seen out there, and they are making hypotheses about what it is. The tails changed significantly between the two sets of Hubble photos, two weeks apart. Based on the information so far, the authors suggest that the tails are all very new, perhaps having been created within the last few months. They even suggest that P5 may be in the process of breaking up, powered by the force of sunlight; the break-up may take a few thousand years. There is a lot of speculation in there, but that's their current model.
Further observations will help, of course. In the meantime, it is at least a striking picture. Don't worry about what it is called. Once again, old simple classifications may be inadequate for the real world. There is more to learn about the asteroid belt.
News stories:
* Hubble Space Telescope spots unprecedented asteroid with six tails. (Nature News, November 8, 2013.) Caution... The lead picture of this story is not a photo, but an "artist's conception". It's very nice, but has little relevance to the object. Don't blame Nature; it's from NASA.
* When is a comet not a comet? (Space Daily, November 11, 2013.)
NASA, central source of information: Hubble site page for P5.
The article: The extraordinary multi-tailed main-belt comet P/2013 P5. (D Jewitt et al, Astrophysical Journal Letters 778:L21, November 20, 2013.) There is a copy of a preprint freely available at the ArXiv: ArXiv copy, preprint.
More from the Hubble Space Telescope:
* Europa is leaking (February 10, 2014).
* A galaxy far, far away: the story of MACS 1149-JD (October 12, 2012).
Other posts about comets and asteroids include...
* The first known Vatira (August 3, 2020).
* Twins? A ducky? Spacecraft may soon be able to tell (August 4, 2014).
* Rings for Chariklo (May 9, 2014).
* Of disasters, asteroids and meteors (February 19, 2013).
* Lutetia: a primordial planetesimal? (February 13, 2012).
* Were comets the source of Earth's water? (February 3, 2012).
* Cometnapping in the stellar nursery (August 4, 2010).
More about tails: An animal that walks on five legs (February 3, 2015).
Previous posts about lawn sprinklers or badminton shuttlecocks: none.
Suggested genes for autism challenged
November 18, 2013
Briefly noted...
A year or so ago an article was published claiming that the authors had found a genetic signature that predicted autism (or autism spectrum disorders). Now we have a new article claiming that the original report is unlikely to be correct. (What is a "genetic signature"? It's a pattern, based on many genes -- 146 genes in this case.)
Such claims and challenges are common in genetics, especially when dealing with genes that cause small effects. Small effects are hard to find, and the results are subject to various biases, including statistical and sampling. I had heard of the original paper of this dispute, but not noted it -- precisely because I am skeptical of such first reports of genes. Until the report is validated, I'm willing to wait. Of course, we always want results to be validated; we have made that point in many Musings posts. But reports of genes with small effects are unusually prone to problems.
News story: Genetic Test for Autism Refuted -- Replication attempt shows that earlier claims about a genetic test for autism were overblown. (The Scientist, October 25, 2013.) A brief and useful overview of the dispute.
The article, which is freely available: Response to 'Predicting the diagnosis of autism spectrum disorder using gene pathway analysis'. (E B Robinson et al, Molecular Psychiatry 19:859, August 2014.) It links, as reference #1, to the article being challenged; that article is also freely available. I caution that trying to follow the arguments in detail is not easy. Most people will prefer to get the gist of the two papers, and the general arguments used to question the original work.
One recent post on autism was about the development of a blood test. As with genetic identification, caution must be exercised when reading reports of such tests. A blood test for autism? (December 11, 2012).
More... Signs of autism in 2-month-old babies (February 7, 2014).
More about autism is on my page Biotechnology in the News (BITN) -- Other topics under Autism. It includes an extensive list of related Musings posts.
Sleep and the brain drain
November 17, 2013
When is the last time you drained your brain?
According to a new article, the correct answer may be "last night".
Biological processes produce waste products -- at the cellular level. Cells need to get rid of their wastes. In higher organisms, this is commonly done via the blood. However, the brain is carefully protected against the blood, by what is called the blood-brain barrier. How do brain cells dispose of their wastes? This has long been a mystery.
A year ago, a group of scientists found that the cerebrospinal fluid (CSF) was the key. It bathed brain cells, and provided a way for cellular waste to exit. This opened up a new way of thinking about waste removal from the brain, but it raised questions of its own. It wasn't at all clear how the CSF established close contact with the brain cells, to allow effective waste removal.
A new article, from the same group of scientists, offers a more complete description of how CSF serves to remove brain waste -- at least in mice. The key finding here is that the process occurs largely during sleep. Brain cells shrink, leaving more space between the cells. That allows the CSF better access.
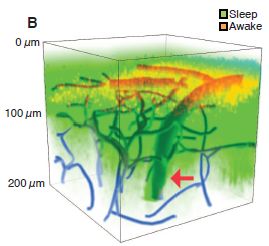
|
This figure gives an idea of what they found.
The general plan was that dyes were injected into the cerebrospinal fluid of a mouse. The distribution of the dyes in the mouse brain was then observed -- by fluorescence microscopy. The green is from a dye that was injected during sleep. The red is from a dye that was injected when the mouse was awake. (Blue shows blood vessels.)
You can see that the dye injected during sleep is observed over a much larger region of the brain.
This is Figure 1B from the article.
|
With this work, we get a better view of how brain waste is removed. The process involves the cerebrospinal fluid bathing the cells. It's done during sleep, when the brain cells shrink a bit, making it easier for the CSF to make close contact with the cells. The pumping of CSF requires considerable energy; doing it during sleep, when the brain is less active, allows it to be done within the energy constraints of the brain.
The study has limitations. For one thing, it is in mice. Perhaps it will hold for other mammals, but that remains to be tested. The article includes an experiment on disposal of proteins, such as the amyloid protein associated with Alzheimer's disease. It's interesting, but is rather artificial; whether it is relevant to the real world remains to be seen. The purpose of noting such limitations is so that we do not overstate what they have done. Their real achievement is substantial. This is pioneering work, which may well lead to a better understanding of the brain, of the brain drain, and of sleep. Pioneering work often gets hyped, and you can see that in the news surrounding this work. The real story is of interest. It is also of interest to note where it might lead, but in doing that we need to be careful to not make claims beyond what has been shown. In this case, it is important to emphasize that nothing here has been shown to apply to humans. So far.
News stories:
* Sleep 'cleans' the brain of toxins. (BBC, October 17, 2013.)
* To sleep, perchance to clean: Study reveals brain 'takes out the trash' while we sleep. (Medical Xpress, October 17, 2013.)
* Two news stories accompanying the article:
1) Neuroscience: Sleep: The Brain's Housekeeper? (E Underwood, Science 342:301, October 18, 2013.)
2) Neuroscience: Sleep It Out. (S Herculano-Houzel, Science 342:316, October 18, 2013.) This includes a nice cartoon figure summarizing the main ideas of the work.
* The article: Sleep Drives Metabolite Clearance from the Adult Brain. (L Xie et al, Science 342:373, October 18, 2013.)
Other posts on sleep and daily rhythms include...
* What happens to capillary blood flow in the brain during sleep -- and why? (November 8, 2021).
* When do jellyfish sleep? (September 29, 2017).
* What if a lion came into your hotel room while you slept? (July 20, 2016).
* Does it matter when you eat? Or whether you leave a light on at night? (December 1, 2010).
* Sleepy teenagers (July 23, 2010).
Two sections of my page Biotechnology in the News (BITN) -- Other topics are relevant here: Brain and Alzheimer's disease. Both include extensive lists of related Musings posts.
Thanks to Borislav for noting this article.
Down syndrome: Could we turn off the extra chromosome?
November 15, 2013
Down syndrome (DS) is a serious human disease caused by a genetic abnormality. DS is caused by having an extra copy of chromosome 21; that condition is called trisomy 21. Chromosome 21 is one of our smallest chromosomes, but it contains a few hundred genes. Exactly which of those genes are responsible for the effects seen in DS is unclear; it is likely that multiple genes are involved.
Is it possible that we could turn off that extra chromosome? A new article offers an approach and demonstrates some encouraging lab results.
The basis for the new work is a perfectly normal and common example of turning off an "extra" chromosome. It's something half of you do all the time. In humans (and in mammals in general), females have two X chromosomes, whereas males have only one. Does this mean that females make twice as much of products coded for by the X chromosome? No. There is a process to turn off the "extra" X chromosome in females. It's called X-chromosome inactivation. It seems to count X chromosomes, and turn off all but one. We know that from people who have more than two, for some reason. The result is clear: everyone has one X active; any X beyond one are inactivated.
How X-inactivation occurs is at least partially understood. It depends on a gene called XIST. XIST codes for an RNA product, which coats the chromosome and prevents its function. An intriguing property of this XIST-based system for chromosome inactivation is that XIST inactivates only the chromosome that makes it. This is the property exploited in the new work.
The key step in the new work was to insert a copy of the XIST gene into chromosome 21. Indeed, the result was that the copy of chromosome 21 with XIST was turned off. Thus, XIST seems to be portable.

|
Here is one of the results showing that turning off one copy of chromosome 21 in DS-derived cells may be useful.
The general plan... They have four cell cultures, and they do a procedure that should induce the cells to differentiate into neural structures. All the cultures were examined 14 days after that induction treatment.
Look at the four pictures. It should be clear that one is quite different, with evidence for structures forming, whereas the other three lack those structures.
|
The one that shows structures is the one from DS cells that have been treated to turn off one copy of chromosome 21. All the others are from DS cells where all the chromosomes are active. This speaks for the success of the procedure.
Some details... The work here uses induced pluripotent stem cells (iPSC) from a DS patient. The two cultures on the left are from the parental DS cells. The two cultures on the right are from a cell line to which the XIST RNA gene has been added; this cell line is called Clone 3. Then there is "dox". What's dox? They have cleverly designed their new cell lines so that they can turn XIST on or off. By default, XIST is off. To turn it on, they add a drug, doxycycline -- or "dox". Note then that Clone 3 makes the neural structures only when dox is added (lower right). Adding dox to the culture of parent cells (lower left) is a control. The observed effect requires both the XIST gene and adding dox to turn it on.
This is Figure 5c from the article. The scale bar (lower right) is 100 µm.
|
The new work shows that it is possible to transplant XIST to a new chromosome, and have it work to inactivate that chromosome. That is a breakthrough. The article also provides some evidence that doing that with one of the extra copies of chromosome 21 in DS cells may be of benefit.
The prospect of treating Down syndrome by turning off the extra chromosome is something for the distant future. In the meantime, the procedure may be useful for research.
News stories:
* Scientists Show Proof-Of-Principle for Silencing Extra Chromosome Responsible for Down Syndrome. (Science Daily, July 17, 2013.)
* Shutting Down the Extra Chromosome in Down's Syndrome Cells. (E Yong, Not Exactly Rocket Science (National Geographic blog), July 17, 2013.) This includes a nice, well-labeled photograph of the chromosomes from a person with DS. (The picture in the Science Daily story is a diagram.)
The article: Translating dosage compensation to trisomy 21. (J Jiang et al, Nature 500:296, August 15, 2013.) Put the article title into Google Scholar, and you will probably find a freely available copy.
More about Down syndrome:
* A hormone treatment that might lead to cognitive improvement in Down syndrome (November 8, 2022).
c
* Why human egg cells become increasingly defective as the mother ages (June 21, 2016).
* Hobbits: Is the answer "Down syndrome"? (December 12, 2014).
* Is it possible that mental retardation could be prevented by a simple prenatal treatment? (January 14, 2013).
More XIST: X-chromosome inactivation in males -- in cancer (February 6, 2023).
The work here made of zinc finger nucleases (ZFN) for targeting the genetic modification. ZFN were discussed in the post Gene therapy: Curing an animal using a ZFN (August 9, 2011).
CRISPR: an overview (February 15, 2015). This includes a complete list of Musings posts on various gene-editing tools, including CRISPR, TALENs and ZFNs.
Developments in making iPSC: Improving the efficiency of making induced pluripotent stem cells (iPSC) (February 1, 2014).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Brain
More about gene therapy is on my Biotechnology in the News (BITN) page Agricultural biotechnology (GM foods) and Gene therapy. We note that the new work might be considered beyond gene therapy; it is chromosome therapy.
There is more on stem cells on my page Biotechnology in the News (BITN) - Cloning and stem cells. It includes a list of related Musings posts.
November 13, 2013
An interesting skull, and a re-think of ancient human variation
November 12, 2013
Look at the following skulls. What do you think? How similar do you think they are? Do you think they might all be the same species?
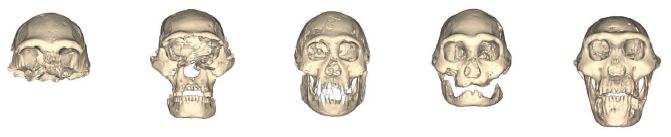
|
This is the top part of Figure 2 from the article.
|
The figure shows five skulls found in Dmanisi, Georgia. They are thought to be about 1.8 million years old -- and they are human, at least in the broad sense.
A new article reports the fifth of these (on the right). It's remarkably complete, one of the best human fossils from that era. That skull #5 is worthy of a paper. In fact, much of the new article is a detailed analysis of that skull.
Then the authors put together the set of skulls. The five skulls were all found in the same area, and they all date to about the same time (to within a few hundred years at most --an instant in geological, or evolutionary, time). Isn't it likely, therefore, that they are all the same species? Yet they are so varied! The authors suggest that what is seen here is the natural variability within that species. That is, they suggest that early humans varied widely -- just as modern humans do.
That suggestion is disturbing to some. There has long been a tendency to subdivide prehistoric humans into multiple species by differences in appearance -- differences that are no greater than those seen here. A problem is that we rarely have multiple fossils from the same time and place; therefore, we have little idea of what the natural variability was within a species of early humans. When somewhat different types of fossils are found from different places and times, there is a natural tendency to call them different species. We know about variability of modern humans; we have not known about the variability within species or populations of early humans -- until now.
There is no easy way to resolve this. The whole notion of species for fossils is difficult. The point of the new article is to put an alternative view on the table. Scientists will now debate these alternative views, looking for evidence that might distinguish them. It will be a lively debate -- and quite possibly without resolution.
Emphasize... The new findings do not show that all early humans in Africa of a given age were a single species. It more raises the question, pointing out that the variability seen among African fossil humans is consistent with them being a smaller number of species than commonly suggested.
That's the story. The new article presents a remarkable new skull from the early history of humans. Beyond that, the authors suggest that variability within early human species was high, and that there may have been fewer species than we have come to accept.
News story:
* Ancient Georgian Ancestors. (The Scientist, October 17, 2013.)
* 1.8M-year-old skull gives glimpse of our evolution, suggests early man was single species. (Phys.org, October 17, 2013.)
* News story accompanying the article: Paleoanthropology: Stunning Skull Gives a Fresh Portrait of Early Humans. (A Gibbons, Science 342:297, October 18, 2013.)
* The article: A Complete Skull from Dmanisi, Georgia, and the Evolutionary Biology of Early Homo. (D Lordkipanidze et al, Science 342:326, October 18, 2013.) Check Google Scholar for a freely available copy.
More about ancient humans:
* Human origins, and the species question (March 28, 2011).
* Did Lucy butcher a cow? (February 11, 2011).
* The Siberian finger: a new human species? -- A follow-up in the story of Denisovan man (January 14, 2011).
More about human skulls:
* Skull surgery: Inca-style (August 21, 2018).
* Who mismeasured man -- and why? (September 9, 2011).
More about re-thinking classifications:
* A new species of orangutan? (January 16, 2018).
* Was Linnaeus's original elephant African or Asian? (December 7, 2013).
Methane on Mars? Follow-up
November 11, 2013
Original post: Cows on Mars? (November 7, 2012)
A year ago we discussed the state of affairs regarding measuring methane on Mars. Briefly, measurements from Earth or from Mars orbit were giving mixed results; some of them suggested that there must be a significant source of methane on Mars. That allows for the possibility that there might be a biological source of methane on Mars. (Although any such source would most likely be microbial, it has become common jargon to use the cow as a unit of methane production for Mars.)
One reason for the review of Martian methane at that time was that NASA had just sent a new lander to Mars -- one with enhanced ability to measure methane. The results are in; in fact, we hinted at them in the earlier post, based on preliminary information. We now have a formal article reporting the many sets of measurements that have been taken over the year. Curiosity finds only traces of methane on Mars; none of the results are significantly different from zero.
The new results do not close the door to low levels of methane, or to methane at other places or times. And they don't close the door to there being life there. However, the fact is, at this point, we have no good evidence for the presence of methane on Mars.
Sorry.
News story: Life on Mars hopes fade after methane findings (Update). (Phys.org, September 19, 2013.)
The article: Low Upper Limit to Methane Abundance on Mars. (C R Webster et al, Science 342:355, October 18, 2013.)
More from Mars: What causes gullies on Mars? (September 8, 2014).
A better way to divide 6 by 2: A more efficient way to use sugar
November 10, 2013
Common sugars, such as glucose, are compounds with six carbon atoms. To focus on just the carbons, we may write them as C6. A key compound in the process of metabolizing sugar is acetic acid, a C2 compound.
Acetic acid is commonly "carried" in the cell by coenzyme A. We refer to this form of acetic acid as acetyl coenzyme A, or acetyl CoA. If it were free, the acetic acid would be acetate ion, because cellular pH is near neutral. Our purpose here is simply to count C, and it doesn't matter whether we have acetic acid, acetate ion, or acetyl-CoA. It's all C2.
The main paragraph above summarizes some key points about how we use sugar -- and raises a question: How many C2 units (acetate) can you get from a C6 unit (sugar)? The answer, more or less universal in biology, is 2. Why only 2? Because the first step in breaking down sugar is to break it into C3 pieces. Each C3 is then broken down to C2 + C1; the C1 is carbon dioxide gas, CO2, and is "lost". The result: each sugar contributes 2 C2 units to further metabolism.
Here is a summary of that, just looking at the carbon count: C6 → 2 C3 → 2 C2 + 2 C1. The C1 units are "lost", as CO2. Thus what an organism effectively gets is C6 → 2 C2. In more mathematical terms, we might say that 6 divided by 2 is 2, with remainder 2 CO2.
This loss of CO2 interests biologists. For example, we grow yeast for the purpose of making ethanol, CH3CH2OH, for use as fuel. One might think that the limit would be making 3 molecules of the C2 compound ethanol from each molecule of glucose. However, the yeast does what we showed above, losing 2 CO2 before getting to 2 C2. Thus the limit effectively is 2 molecules of ethanol per molecule of glucose -- 2 C2 per C6.
A team of biochemical engineers has now addressed this problem -- and re-engineered the basic metabolism of a common bacterium to avoid that initial loss of CO2.
The following figure shows an example of what they have accomplished. Caution... The figure is a little different from what the above discussion might lead you to expect, and the controls are not easy to explain. This is a complex biological system, and they have made multiple changes.
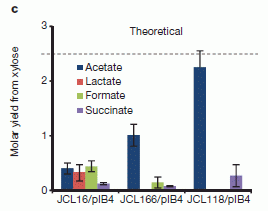
|
In this test, they use xylose, a C5 sugar. Just counting carbons, the theoretical yield of acetate, C2, would be 2.5 acetate per xylose. That theoretical yield is shown with a dashed line in the figure. (In fact, there are numerous problems with growth on xylose, and they have dealt with various of them here.)
They test three strains, but we will look only at one of them: the strain on the right (with the complicated name (JCL118/pIB4).
|
You can see that they obtained a yield of about 2.3 molecules (or moles) of acetate per molecule (or mole) of xylose. That's better than any other result shown, and is approaching the theoretical yield of 2.5. (The "old" pathway, involving a loss of CO2 from C3 units, would give an expected yield of 1.67 from C5 xylose; they seem to be well above that.)
This is Figure 4c from the article.
|
This work marks a major development. The scientists have succeeded in making a major change in basic metabolic pathways. Given how complicated and interconnected those pathways are, that is hard to do. It's also fair to say that it remains to be seen how well these engineered strains will do when grown for a long time. Despite that reservation, this work seems to have improved the efficiency of how a microbe converts a common sugar to a useful product. In simple terms, they have gone from making 2 C2 to making 3 C2 per C6; that is a 50% increase. It will be interesting to see how this development is followed up.
The authors call their new metabolic process non-oxidative glycolysis, or NOG. The name notes its relationship to the common process of glycolysis, but now without the loss of CO2, an oxidative step. The new process can be seen as a substitute for glycolysis, a better way to get from C6 to C2; however, it is not a simple modification, but more of a complete reworking.
News story: UCLA engineers develop new metabolic pathway for more efficient conversion of glucose into biofuels; possible 50% increase in biorefinery yield. (Green Car Congress, October 1, 2013.)
The article: Synthetic non-oxidative glycolysis enables complete carbon conservation. (I W Bogorad et al, Nature 502:693, October 31, 2013.)
Another example of trying to engineer genetic improvements to the efficiency -- and economics -- of making ethanol: Engineering E coli bacteria to convert cellulose to biofuel (December 13, 2011).
That work is from the same people who worked out a process for making the anti-malaria drug artemisinin in microbes. That project involved extensive genetic engineering to promote the flow of C from the feedstock to the desired drug. The newer work, to make fuel, is an extension of that work. The artemisinin work is noted on my page Internet Resources for Organic and Biochemistry under Alkenes.
A post that notes the interconversion of sugars and the connection to glycolysis: Fructose; soft drinks vs fruit juices (November 7, 2010).
My page Organic/Biochemistry Internet resources has sections on Carbohydrates and Energy resources. Each includes a list of related Musings posts.
DNA twisting and long genes -- and autism: Are there connections?
November 8, 2013
Odd story. Intriguing story. The heart of the story is that genes involved in autism may be long genes. Long genes? Why would gene length be an issue, and how can one tell?
The story is tentative, but involves some interesting issues about DNA. As you know, ordinary DNA has two strands. They are wound around each other. Using DNA, for example to make messenger RNA from it, requires that the DNA be unwound, so that the "inside" of the coding bases can be seen. Unwinding DNA is not easy. You can't just pull the two strands apart; that results in a tangled mess -- not unlike what happens with a phone cord or such that gets used a lot.
Cells contain enzymes that help with unwinding DNA. What do they do? What reactions do they catalyze? One class of enzyme simply changes the winding of the DNA. For example, the enzyme may cut the DNA, twist one end around the chain, and repair the cut. The result? DNA that has one less twist. These enzymes are known as topoisomerases; they isomerize the topology of the DNA, in this case, the twist. Some antibiotics work by inhibiting a bacterial topoisomerase, attesting to the importance of such enzymes.
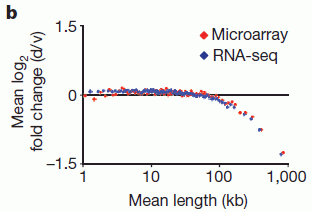
|
This graph shows the effect (y-axis) of inhibiting topoisomerase vs gene length (x-axis).
The curve starts at about zero (no effect), for short genes. Then, for long genes, the curve goes negative. "Negative" here means that inhibiting the topoisomerase reduces gene expression. That's the key message from this graph.
This experiment was done with mouse neurons in cell culture. Additional work suggests that the main result holds broadly with mammalian cells, including human neurons.
|
For those who want the fine print...
Two methods for measuring RNA were used. These are shown by the two colors of data points; the two methods gave the same results.
The y-axis, which shows the response, plots the ratio of gene expression with drug to that without the drug. That ratio is called d/v, where d = drug, and v = vehicle -- a control using simply the solvent (or "vehicle") for the drug. The d/v ratio is shown on a log2 scale. The final values, at about -1.5 on the graph, have d/v approximately = 1/3. The ratio shown is the average for all genes of that gene length. The x-axis is also on a log scale: log10.
This is Figure 1b from the article.
|
The finding here, as the graph shows, is that the function of long genes is particularly sensitive to the level of DNA topoisomerases. Apparently, unwinding long genes so that they can be transcribed ("read") is a problem. That is "logical", but there had not been any good evidence that the problem is significant.
So, what is the connection to autism? There is nothing in the experiment above about autism -- other than that they used neurons. However, people have noticed that the list of genes implicated in autism seems to include many long genes. Further, among the genes implicated in autism are genes for topoisomerases. Most identification of genes for autism is tentative, but it is intriguing that mutations that reduce the function of an enzyme involved in unwinding DNA might be associated with autism. The current experiment helps reinforce that connection.
Where will this lead? I think the best approach for now is to consider this "basic" science. We want to know more about genes and about the disease process. As we learn more, we can then see where it leads.
News story: A Potential Cause of Autism? Key Enzymes Are Found to Have a 'Profound Effect' Across Dozens of Genes Linked to Autism. (Science Daily, August 28, 2013.)
* News story accompanying the article: Autism: A long genetic explanation. (R N Plasschaert & M S Bartolomei, Nature 501:36, September 5, 2013.)
* The article: Topoisomerases facilitate transcription of long genes linked to autism. (I F King et al, Nature 501:58, September 5, 2013.)
More about twisted DNA... DNA: Watching the hopping supercoils (November 24, 2012).
Previous post on autism... A blood test for autism? (December 11, 2012).
More about autism is on my page Biotechnology in the News (BITN) -- Other topics under Autism. It includes an extensive list of related Musings posts.
November 6, 2013
Slavery - II
November 5, 2013
A previous post noted an article on slavery: Slavery (January 22, 2010). The gist of the article was that more people are enslaved now than at any earlier time in human history.
We now have a follow-up -- a major follow-up. A new organization, formed for the purpose, has issued a report on slavery worldwide. The report offers numbers, but -- more importantly -- reasons. Those who have studied American history will recall that slavery was deeply embedded in some parts of American culture. Slavery is still embedded in cultural practices around the world. The important part of the report is the discussion of these practices, and what is being done about them.
The report will be an annual publication, according to plan. Tracking progress is the goal. This 2013 report is simply a baseline.
News story: More Than 29 Million People Live As Slaves, According To New Report. (Huffington Post, October 17, 2013.)
The report: Global Slavery Index. (Walk Free Foundation, 2013. Updated annually.)
From the 2013 report (original post)...
The United States ranks #134 (out of 162 countries studied) on their list, by prevalence of slavery. They estimate about 60,000 are enslaved in the US, out of a population of 314 million; that is about 0.02%. US slavery is largely due to trafficking.
Prevalences, on a percentage basis, range from 4.0% (#1) down to 0.007% (three countries tied for #160).
My page for Biotechnology in the News (BITN) -- Other topics includes a section on Ethical and social issues.
Bees: Why pollen might be bad for them
November 4, 2013
Briefly noted...
In previous posts we have noted why pollen may be good for bees [links at the end]. A new article is a caution: it shows why pollen might be bad for bees.
The concern is that pollen may carry poisons, such as fungicides sprayed on the crop. The scientists show that pollen may contain such poisons, and then they show that indeed the bees may be affected by such pollen.
Neither of those results is particularly surprising, but now we have data. The story of bees and pollen is complicated.
Is there a solution to this new problem? The authors note that certain kinds of spraying of crops is withheld when it might affect pollinating bees. Fungicides are not currently included in the list of suspect agents. Maybe they should be.
News story: Common agricultural chemicals shown to impair honey bees' health. (EurekAlert!, July 24, 2013.)
The article, which is freely available: Crop Pollination Exposes Honey Bees to Pesticides Which Alters Their Susceptibility to the Gut Pathogen Nosema ceranae. (J S Pettis et al, PLoS ONE 8(7):e70182, July 24, 2013.)
Background posts on bees and dietary pollen:
* Should bees eat honey? (July 12, 2013).
* Why are the bees dying? (January 26, 2010).
More on bees: A plant virus that grows in bees: role in colony collapse? (February 17, 2014).
Thanks to Borislav for noting this article.
Drug degradation: A drug that reappears after being degraded
November 2, 2013
An important issue is the fate of chemicals in the environment. Of particular interest is man-made chemicals, sometimes called foreign chemicals. For example, if a drug is developed, some of it ends up in the environment, either after going through us or simply as waste from the container. What happens to it, and what are the implications?
A new article is a reminder that the results may be more complicated than they seem at first.
The general idea of the article is that the scientists study a particular drug. It is known that the drug is inactivated by light; that is "good". The particular drug is used for farm animals. Being inactivated by light is relevant to its release in the field. However, what they find is that this change is reversible. At night, the inactivated drug changes back to the original drug!
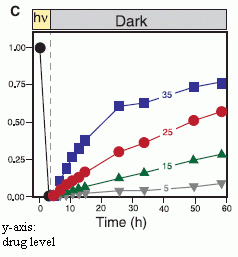
|
Here is an example of their results.
The graph shows the concentration of one form of the drug, one that is bioactive (and therefore of concern), over time.
The y-axis scale is relative concentration, with "1.00" being the starting level. Time is in hours; importantly, the lights were on for the first 12 hours (labeled "hν"), and then were turned off (labeled "dark" -- an artificially long night for this experiment.)
|
What does that "hν", at the top, mean? The ν is the Greek letter nu. ν is the symbol for the frequency of the light (related to wavelength), and h is Planck's constant. hν is the energy of light of a certain frequency. Interestingly, as here, scientists sometimes use hν as a general symbol for light energy.
This is Figure 2C from the article. (I added a label for the y-axis.)
|
So, what do we see? During the light phase, with hν, the drug concentration declines, more or less to zero. Then, darkness. The drug reappears. There are four sets of data -- at various temperatures (T), as shown by the numbers (°C) on the lines. The higher the T, the faster the drug returns; that's expected from ordinary chemistry.
Yes, the time scale is long for the dark phase. They do this here to make the effect clear; there is significant reactivation within 12 hours. However, they also note that extended darkness may be relevant to that portion of the drug that binds to something and sinks.
Thus we see that a drug which seems to have disappeared can reappear. The degradation is a more complex issue than we thought. This needs to be studied further.
For those who would like some of the chemistry behind this... Here is Figure 3 [link opens in new window] from the article, with their legend. The starting compound, 17α-trenbolone (17α-TBOH), is shown at the upper left. For this work, the key part of the molecule is the system of conjugated double bonds, shown in red; it starts with the O at C3 (lower left) and weaves it way up to C12.
In the light-induced reaction, water is added across the double bond system. For the major product, the -OH is added at position 12, and the H is added at position 4. This gives 12-hydroxy-17α-trenbolone, shown at the upper right. The lower part of the figure shows the reversal of this reaction.
News story: Steroids may persist longer in the environment than expected. (Phys.org, September 26, 2013.)
* News story in the journal (earlier issue, when the article was originally posted online prior to publication): Ecology: Zombie Endocrine Disruptors May Threaten Aquatic Life. (E Stokstad, Science 341:1441, September 27, 2013.)
* The article: Product-to-Parent Reversion of Trenbolone: Unrecognized Risks for Endocrine Disruption. (S Qu et al, Science 342:347, October 18, 2013.)
Here are a few other posts about degradation issues. Each links to more.
* How to dispose of unused medicines (September 10, 2012).
* Degradable polyethylene isn't (October 17, 2011).
* Developing improved degradation of organophosphate pesticides (September 7, 2010).
Atomic bombs and growing new brain cells
November 1, 2013
We recently noted the story of how open-air testing of atomic bombs in the 1950s caused a spike in the level of radioactive carbon, C-14, in the atmosphere, and how this effect could be used for dating [link at the end]. Here we have another example of using the same basic phenomenon. In this case, the scientists are dating human brain cells. The question is whether or not humans make new brain cells as adults. The scientists look at the C-14 in DNA, a relatively stable molecule.
If humans do not make new brain cells as adults, then the C-14 level in the brain DNA will correspond to their birth year. However, if they make new brain cells, then the C-14 level will appear to be from more recent times. Importantly, the scientists look at specific types of brain cells; it has long been suspected that the answer may differ for different parts of the brain. (That's been shown for mice.)
|
This figure shows the basic finding.
The graph shows the amount of C-14 in the hippocampal DNA (y-axis) from people born in various years (x-axis). The analyses were all done in the decade of the 2000s, using autopsy samples. The solid line shows the calibration curve, which is similar to that shown in the background post.
This is Figure 3 from the article.
|
The key observation is that the amount of C-14 found always corresponds to a later time than the birth year. The simple interpretation is that some of the hippocampal cell DNA was made after the birth year.
More specifically, the amount of C-14 found is above the calibration line for the older people (those born prior to the peak region). This means that all of them have made hippocampal DNA since about 1955. Further, none of them have C-14 levels as high as the later C-14 levels; this means that any such new hippocampal DNA is only partial.
There is an important control, not shown here. In earlier work, they showed that the C-14 level was close to the calibration curve for some other brain regions. That is, the results here, for hippocampal cells, stand out as special. The hippocampus is special: it's a region for memory formation and learning. In rodents, there is good evidence that new hippocampal neurons are important for memory.
They do extensive mathematical modeling, and come up with their best estimate of what is going on. Whether they have that all correct is open for further work. The main point is that they seem to have good evidence, based on using the C-14 from the era of open testing of atomic bombs, that humans do make hippocampal neurons throughout life.
News stories:
* Nuclear Testing from the 1960s Helps Scientist Determine Whether Adult Brains Generate New Neurons. (Science Daily, June 6, 2013.)
* Human Adult Neurogenesis Revealed. (The Scientist, June 7, 2013.)
* Weapons testing data determines brain makes new neurons into adulthood. (Lawrence Livermore National Laboratory, June 10, 2013.) From one of the participating labs.
* News story in Science: Neuroscience: What the Bomb Said About the Brain. (G Kempermann, Science 340:1180, June 7, 2013.)
* News story accompanying the article: (Radio)active Neurogenesis in the Human Hippocampus. (M A Kheirbek & R Hen, Cell 153:1183, June 6, 2013.)
* The article: Dynamics of Hippocampal Neurogenesis in Adult Humans. (K L Spalding et al, Cell 153:1219, June 6, 2013.) Check Google Scholar for a freely available copy.
Background post: Atomic bombs and elephant poaching (October 25, 2013).
More about atomic bombs: Analysis of uranium samples from World War II Germany (November 7, 2015).
More about making new neurons: Formation of new neurons in adults: relevance to Alzheimer's disease? (May 21, 2019)
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain. It includes a list of brain-related Musings posts.
October 30, 2013
When does global warming occur: day or night?
October 28, 2013
Briefly noted...
The purpose here is to note an idea behind a recent paper. It's broadly accepted now that the Earth's temperature (T) has been increasing over recent decades. Of course, that increase is not simple. It varies from year to year, and it varies by location. The new article builds on a point that is not often mentioned: warming varies by time of day -- and this might be important.
The climate records show clear evidence for this variation. Overall, it seems that the nighttime T increase is about 40% greater than the daytime T increase.
Why does it matter? An important issue in making predictions about climate change is dealing with how plants respond. Plants remove carbon dioxide, CO2, from the air by photosynthesis; they are likely to do so more as the CO2 level and T both increase. Thus plants may moderate the effects of CO2 increase. (That's why clearing forests is bad.) However, plants grow differently day vs night. They photosynthesize only during the day; they respire at all times, including at night. It's a reasonable first guess that increasing T affects both processes similarly. If the T increase is higher at night, then the effect on nighttime respiration may be larger than the effect on daytime photosynthesis. Therefore, the ability of plants to reduce atmospheric CO2 may be less than we had expected.
The article explores this, and comes up with a mix of results. Some fit the model suggested above, some do not; it may be that the details matter. I don't want to get into the results for now, but would rather just note the idea, which deserves attention. This article is a start.
News story: Effects of uneven warming on Northern Hemisphere vegetation assessed. (The Arctic, September 6, 2013.) This item is no longer available, and is not at the Internet Archive. I have not found a good replacement item. The original URL was: http://arctic.ru/news/2013/09/effects-uneven-warming-northern-hemisphere-vegetation-assessed
* News story accompanying the article: Biogeochemistry: As different as night and day. (C Still, Nature 501:39, September 5, 2013.)
* The article: Asymmetric effects of daytime and night-time warming on Northern Hemisphere vegetation. (S Peng et al, Nature 501:88, September 5, 2013.) Check Google Scholar for a freely available copy.
More about global warming:
* National contributions to global warming (June 25, 2014).
* Why the lull in global warming? (February 11, 2014). This focuses on another explanation for the current lack of rising T.
* SO2 reduces global warming; where does it come from? (April 9, 2013).
* Global warming trend? Independent evidence (March 22, 2013).
More about CO2: Can a coral adapt to a more acidic ocean? (September 29, 2013).
More about photosynthesis: Photosynthesis that gave off manganese dioxide? (July 21, 2013).
A new type of dengue virus
October 27, 2013
Dengue virus is a serious human pathogen, with an expanding range. A special problem with dengue is that there are four types of dengue virus -- and they interact in a very troublesome way. Infection with one type leads to good immunity against that type of virus, but increases the risk of serious disease upon infection with another dengue type.
It is as if antibodies against one type enhance infection with another type. In fact, there is some evidence for that. Whatever the details behind this, it has implications for design of a dengue vaccine. It is reasonable to worry that a vaccine against dengue could enhance infection.
We now have an addition to the dengue story: the discovery of a fifth type of dengue virus. The implications for the bigger story are not known; there is no information yet on how this new type interacts with the other types. All we have so far is an announcement, based on a talk presented at a meeting last week. We just briefly note, then, the announcement of a fifth type of dengue virus -- with unknown implications.
News story: First New Dengue Virus Type in 50 Years. (Science Insider, October 21, 2013.)
Other posts about dengue include:
* Dengue vaccine follow-up: Phase 3 trial (September 15, 2014).
* A dengue vaccine trial (December 1, 2012).
* Dengue fever: an overview (February 28, 2011).
* Dengue fever -- Two strikes and you're out (August 10, 2010). This post notes the discovery of anti-dengue antibodies that seem to help other dengue types infect.
Atomic bombs and elephant poaching
October 25, 2013
Carbon-14 (C-14 or 14C) is a radioactive form (isotope) of carbon. Because it decays at a known rate, it can be used for dating. C-14 dating assumes that C-14 is incorporated into an organism during life. After death, there is no further incorporation, but there is decay. If we know the original amount of C-14 incorporated, we can calculate the age by measuring how much is left. The amount of C-14 is measured relative to the amounts of the other carbon isotopes, which are stable.
Knowing how much C-14 was present originally is not a trivial issue. Much work has been done trying to sort out that part of the story; sometimes, simple assumptions are made.
There is an interesting special case regarding the level of C-14 in the atmosphere (the general source of C-14 for organisms). During the 1950s, atomic bombs were tested in the open. This resulted in the release of substantial amounts of C-14 to the atmosphere. Biologists have taken advantage of this burst of C-14 to do special types of dating experiments. A recent paper gives an interesting example.
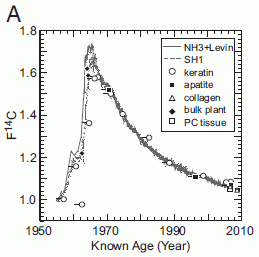
|
The graph shows their calibration. The graph plots the C-14 content (y-axis) vs the known age of the samples (x-axis).
The C-14 content is expressed here as something they call F14C, or fraction modern carbon. It's based on measuring the fraction of the carbon that is C-14.
You can see that there is a clear pattern of C-14 content vs year. Further, that pattern shows a burst of C-14 in the 1950s.
|
A complication is that the curve shows a peak; simply knowing the F14C allows either of two dates to be assigned, one on the rising part of the curve and one on the falling part. For example, an F14C value of 1.2 could mean that the sample was from either 1960 or 1990. Further information would be needed to distinguish those two possibilities.
This is Figure 1A from the article.
|
The general idea of the curve above is not new. The authors claim theirs is the best C-14 calibration curve yet made for this time span.
One reason this article has gotten attention is because of a proposed application. Among the samples the authors studied are teeth, tusks, and horn from herbivores -- such as elephants. They show that such samples can be dated to within about a year, for samples dating since 1955, that is, for samples on the recent side of the curve. They thus suggest that such C-14 dating may be helpful in wildlife forensics issues, such as elephant poaching. Precise dating of the material, such as tusks, is important, but it has been difficult.
News stories:
* Carbon from nuclear tests could help fight poachers. (BBC, July 1, 2013.)
* Nuke Test Radiation Can Fight Poachers Who Kill Elephants, Rhinos, Hippos. (Science Daily, July 1, 2013.)
The article: Bomb-curve radiocarbon measurement of recent biologic tissues and applications to wildlife forensics and stable isotope (paleo)ecology. (K T Uno et al, PNAS 110:11736, July 16, 2013.)
Follow-up: Carbon-14 dating of confiscated ivory: what does it tell us about elephant poaching? (February 10, 2017).
Another application of the same C-14 curve: Atomic bombs and growing new brain cells (November 1, 2013).
More about C-14 dating:
* Eye analysis: a 400-year-old shark (September 3, 2016).
* Tree rings, carbon-14, cosmic rays, and a red crucifix (July 16, 2012).
My page of Introductory Chemistry Internet resources includes a section on Nucleosynthesis; astrochemistry; nuclear energy; radioactivity. That section links to Musings posts on related topics, including the use of radioactive isotopes.
Other posts about elephants include:
* Was Linnaeus's original elephant African or Asian? (December 7, 2013).
* If the elephant can't find its dinner, should you help by pointing to it? (October 18, 2013).
More about horns... Polled cattle -- by gene editing (July 8, 2016).
More forensic science: How easy is it to destroy any traces of 43 students by burning them? (October 25, 2016).
October 23, 2013
Are there genetic issues that we don't want to know about?
October 22, 2013
We are in an era of unprecedented ability to study genes -- of an individual or of the human population as a whole. Is this good? One would think so; knowledge is good. But new knowledge can also raise questions. The journal Nature has recently published a news feature about the possibility that there are genetic issues we don't -- as a human society -- want to know about. One example is the genetics of intelligence.
I encourage you to browse it. It presents some good questions and examples. It doesn't really break any new ground; in that sense, I was disappointed with it. But that's ok. It's a useful summary, with four candidate issues discussed together.
Various privacy issues have been in the news recently. The issues here might be thought of as privacy issues.
If you find yourself confused by all this, that is good. If you find that you have different feelings about different privacy issues, ok. The point is to be aware of these issues -- and how complicated they are. These are real issues. Don't look to the article (or to me) for answers. I'm not sure there are any.
News feature, which is freely available: Ethics: Taboo genetics. Probing the biological basis of certain traits ignites controversy. But some scientists choose to cross the red line anyway. (E C Hayden, Nature 502:26, October 3, 2013.)
There have been many Musings posts about the use of genetic information. In particular, an early post on personalized medicine includes an extensive list of related posts, some of which are relevant here. Personalized medicine: Getting your genes checked (October 27, 2009).
There is more on personalized medicine, including useful background and perspective, on my page Biotechnology in the News (BITN) - DNA and the genome. Especially see the sections "Examples of how genome information is useful" and "Recent items, briefly noted". There is also as section of related Musings posts.
Also see:
* Identifying individuals from their genomes: a controversy (December 5, 2017).
* Pangenomes and reference genomes: insight into the nature of species (February 7, 2017).
How a cork causes an off-flavor in a beverage
October 21, 2013
Briefly noted...
Off-flavor can be an issue with foods and beverages. (Flavor includes what we would commonly call both taste and odor.) An example is cork taint of wines, where the wine seems to give a sensation of something from the cork. In fact, one chemical has been associated with cork taint: 2,4,6,-trichloroanisole (TCA).
|
2,4,6,-trichloroanisole (TCA).
This is part of Figure 1F from the article.
|
It has generally been assumed that the odor of TCA is responsible for the cork taint. However, there has never been any direct demonstration of that. A new article offers some evidence for a different interpretation: that TCA is an inhibitor of ordinary odor responses, rather than an odor itself.
The new article provides evidence for this by direct testing of neuron responses. The results show that TCA inhibits odor neurons at extremely low concentrations. Such inhibition could cause off-flavor either by overall inhibition of odor detection or by distortion of complex odors.
This is an intriguing result. However, there are problems raised by the conclusion; the authors discuss some of them. For example, TCA does have an odor of its own -- an unpleasant odor. Further, the sensitivity to TCA is remarkable; the authors suggest that its ability to dissolve in lipid membranes is behind this, and they provide some interesting evidence, based on comparing chemicals that differ in their hydrophobic character. These are incomplete stories; the authors' suggestions are not entirely satisfying at this point.
I'll leave it at this for now. The experiments are complex. The initial work is done with neurons from amphibians; it's likely it applies to humans, but we don't know that. The work may have uncovered an interesting effect at the cellular level. How this plays out in the bigger story of flavor perception is open for further work.
News story: Corked Wine Plugs Up Your Nose. (Science Now, September 16, 2013.) Interesting picture.
The article, which is freely available: 2,4,6-Trichloroanisole is a potent suppressor of olfactory signal transduction. (H Takeuchi et al, PNAS 110:16235, October 1, 2013.)
More about taste and odor, and such:
* Added August 6, 2025.
Could you tame hot peppers by adding an anti-hot substance? (August 6, 2025). This may be another example of sensory modulation, where an added substance affects the response to another, but has no effect alone.
* Some shrimp in your wine? (August 27, 2016).
* Why and how some cockroaches avoid glucose (October 11, 2013).
* The chemistry of a tasty tomato (June 18, 2012). Includes the notion that flavor = taste + odor.
* What does blue light smell like? (July 18, 2010).
Another anisole derivative: Understanding locust swarming: a chemical to deal with the problem? (October 13, 2020).
A vaccine against malaria -- with 100% efficacy?
October 20, 2013
A new article reports... A new vaccine was given to six people, who were then challenged with malaria. None became infected.
Now, that's a Phase I trial, and six people aren't very many. Nevertheless, the result is at least intriguing. After all, the best malaria vaccine so far, one that might gain official approval, is about 50% effective. Further, there is an interesting story behind this new vaccine, so let's look.
The story of malaria vaccines is one of frustration, as hinted above. Most modern work is based on using individual proteins of the malaria parasite to induce antibodies. It works, but not very well. However, we actually know a good way to induce antibodies against malaria: infect with the parasite itself. It's very good at inducing antibodies. Unfortunately, it also induces disease. There is a solution to that dilemma -- and it has been known for many years. The approach has been used for many vaccines -- dating back to Pasteur. Kill or weaken the parasites. Infection with "attenuated" malaria parasites leads to an immune response, and no disease.
So why not? The problem is practical. How do you do this infection with attenuated parasites? One method was worked out some time ago. Irradiate the mosquitoes carrying the parasite. Irradiate them enough to weaken the parasites, but leave the mosquitoes happy -- happy enough that they will still bite you, and inject their load of parasites. A thousand or so mosquito bites -- bites by mosquitoes carrying irradiated parasites -- should induce good immunity. Yes, practical problems.
There was a recent important development that gave this approach a new life. Scientists learned how to inject malaria parasites directly into people. That's not a trivial problem, but it seems to have been solved. We noted part of that story in an earlier post [link at the end] -- and we hinted it might be relevant to vaccine development.
So we now have a new candidate malaria vaccine, based on injecting irradiated parasites directly into people -- into their bloodstream. It has just undergone a phase I clinical trial, the first level of testing in humans. The results, published last month, showed that the effectiveness increases with dose -- a good sign that the results are biologically meaningful. At the highest dose tested, it was 100% effective. 6 people out of 6. That's what the start of this post refers to.
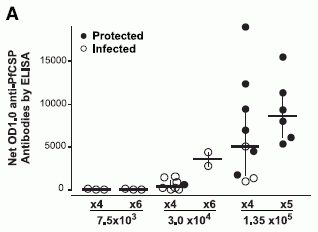
|
This figure summarizes some of the results.
The general layout of the graph is that amount of antibody made is shown on the y-axis vs vaccine dose on the x-axis. Each point is for one person who received vaccine.
Further, each vaccine recipient was "challenged" with malaria at some point after the last vaccine dose. That is, they were experimentally infected. The symbol used for each person's data point shows whether they became infected (open symbols) or were protected (filled symbols).
|
We'll fill in some details in a moment, but here are the main observations...
* There is a general trend that, as the vaccine dose increased, the amount of antibody made increased.
* There is a general trend that, as the vaccine dose increased, the person was more likely to be protected from the challenge infection.
That is, there is a general trend that the vaccine showed increasing effectiveness with increasing dose. Further, protection correlated with measurable antibodies.
More specifically, all persons with the highest vaccine dose were protected; that is the basis for the "6 of 6" claim. They all had high antibody levels. The next highest dose showed protection in 6 of 9, with a general (but imperfect) correlation with antibody level. The lower doses gave low antibody levels, and only rare protection.
Some detail...
We noted that the x-axis shows increasing vaccine dose. However, "dose" is a complicated variable. One part of dose is the number of parasites injected each time; this number is shown (bottom). Another part of dose is how many injections, each at that level, were given. This is also shown (with x and a number). For example, the highest dose shown (right side) is x5 at 1.35x105; this means that the people at this dose received five injections, each with 1.35x105 parasites.
In addition, the timing of the injections may be important, as well as the time between the last dose and the experimental infection. These timing variables are given and discussed in the paper. It is a simplification to refer to increasing dose as we have done here.
This is Figure 3A from the article.
|
That's it. A very encouraging result. It's also a very small, Phase I, study. There will be more.
News story:
* Malaria: 100% protection in early vaccine trial. (Medical News Today, August 9, 2013.)
* News story accompanying the article: Immunology: Pasteur Approach to a Malaria Vaccine May Take the Lead. (M F Good, Science 341:1352, September 20, 2013.)
* The article: Protection Against Malaria by Intravenous Immunization with a Nonreplicating Sporozoite Vaccine. (R A Seder et al, Science 341:1359, September 20, 2013.)
The vaccine was developed by a small company called Sanaria. Their employees are among the authors here.
Background post: An easier way to get infected with malaria (January 18, 2013).
Follow-up: A highly effective malaria vaccine -- follow-up (May 3, 2017).
* Previous post on malaria: Malaria-infected mosquitoes have greater attraction for people (May 28, 2013).
* Next: Pop goes the hemozoin: the bubble test for malaria (January 24, 2014).
There is more about malaria on my page Biotechnology in the News (BITN) -- Other topics under Malaria. It includes a list of Musings posts on malaria.
Another story of a vaccine which tests as 100% effective: An Ebola vaccine: 100% effective? (August 7, 2015).
If the elephant can't find its dinner, should you help by pointing to it?
October 18, 2013
Apparently, according to a new article.
In the new work, scientists tested elephants to see how they respond to the human gesture of pointing. The basic experimental design was that food was in one of two buckets, and the human experimenter pointed to the correct bucket. What did the elephant do?

|
Elephant goes to the bucket that the human points to.
This gives an idea of the testing arrangement, but I encourage you to check out some of the movie files, listed below.
This is Figure 1 from the article.
|
The article gives the overall test results, but also scores for the individual elephants. On the basic pointing test, elephant Coco was correct 46 out of 52 tries. In contrast, elephant Izibulo scored 6 out of 18. Interesting.
Work such as this is interesting in that it begins to reveal the behavioral and communication skills of other animals. Taken at face value, the article suggests that elephants have an innate understanding of the human pointing gesture; no other animal has shown this.
The work raises many questions. Some are addressed by the authors. For example, they do further testing to see whether the elephants are responding to where the person points or to where the person looks. (It's the pointing.) Among other questions, which don't get answered here... The elephants tested here have all lived in captivity and have long been around humans. Does that matter? And intriguingly, do elephants themselves point? The news stories discuss such questions at length. This is one of those articles that opens up a topic we might not have thought to consider; it's not at all clear how close they have gotten to the full story.
News stories:
* Elephants 'understand human gesture'. (BBC, October 10, 2013.)
* African Elephants Understand Human Gestures -- African elephants correctly interpret human pointing cues to find hidden food, without being trained to do so. (National Geographic, October 10, 2013. Now archived.)
Movies. There are short movie files illustrating the tests. Some are about 10 seconds, with a single test. They do not have sound. Each of the news stories has a movie file, and four are included as Supplementary material with the article.
The article: African Elephants Can Use Human Pointing Cues to Find Hidden Food. (A F Smet & R W Byrne, Current Biology 23:2033, October 21, 2013.)
More on elephants and such...
* Was Linnaeus's original elephant African or Asian? (December 7, 2013).
* Atomic bombs and elephant poaching (October 25, 2013).
* Early American art: a 13,000 year old drawing of a mammoth (July 18, 2011).
* Why a tree cultivates ants (October 3, 2010).
Another dinner story... Using a pH meter to help you find dinner (July 8, 2014).
Among other posts on animal behavior:
* Use of instructional videos -- in the wild (November 3, 2014).
* Pink corn or blue? How do the monkeys decide? (June 9, 2013).
October 16, 2013
What to do with old floppy disk drives
October 15, 2013
Check out a couple of short videos at YouTube...
* a duet. (2 minutes.)
* a quartet. (4 minutes.)
As we discussed this prior to posting, I jokingly suggested that this was how one made music with computers prior to MIDI. I was promptly corrected. A reader recognized that "The controller board components are too small to be pre-MIDI". A quick check also showed that MIDI dates back to the early 1980s; the 3.5 inch drive to the mid 80s.
The following Musings post also includes the two pieces featured here: Tesla coils -- music (May 31, 2009).
There is more about music on my page Internet resources: Miscellaneous in the section Art & Music. It includes a listing of music-related Musings posts.
Thanks to various people for contributions and comments.
Did the earliest dinosaurs like flowers?
October 14, 2013
According to conventional wisdom, that is a dumb question. There were no flowers around when the first dinosaurs appeared. Flowers came a hundred million years later.
The following picture, from a new article, says that the conventional wisdom may be wrong.

|
What is it? It's a pollen grain, as seen with an ordinary light microscope. The scale bar is 10 micrometers.
Detailed analysis of this pollen grain makes it clear that it is very similar to pollen from modern angiosperms -- the flowering plants.
This is #4 from Plate I from the article.
|
One more point about this pollen grain... It is about 240 million years old. It's about a hundred million years older than any other accepted evidence for angiosperms.
The new article reports not just this pollen grain, but several, of different types -- all about the same age. The authors use a variety of high resolution imaging methods; the article is full of beautiful pictures.
These angiosperm-like pollen grains date back to the early days of the dinosaurs. However, the authors note that the interpretation is not entirely clear. Are these from true angiosperms -- from plants with flowers? Or are they from plants on an ancestral line destined to yield flowers? We can't tell at this point. What the new results do is to open the door... we now have angiosperms, or plants which appear to be on an angiosperm-like lineage, dating back 240 million years.
We have no idea what the early dinosaurs thought of these plants, whatever they were. However, it is now proper to think about that: we now have evidence that the early dinosaurs co-existed with plants of the angiosperm lineage. More importantly, the new work pushes the time scale of the angiosperms back a hundred million years.
News story: Prehistoric Pollen -- Scientists discover fossilized remnants of a flowering plant about 100 million years older than the oldest previously found. (The Scientist, October 2, 2013.)
The article, which is freely available: Angiosperm-like pollen and Afropollis from the Middle Triassic (Anisian) of the Germanic Basin (Northern Switzerland). (P A Hochuli and S Feist-Burkhardt, Frontiers in Plant Science -- Plant Evolution and Development 4:344, October 2013.)
More about pollen: Should bees eat honey? (July 12, 2013).
More about old flowers: A 30,000 year-old plant, with an assist from a squirrel (March 10, 2012).
More about flowers...
* Why growing sunflowers face the east each morning (November 8, 2016).
* pH and the color of petunias (March 26, 2014).
More about angiosperms: Plants and climate change (April 25, 2010).
* Previous post about dinosaurs: Microraptor was piscivorous (May 25, 2013).
* Next: How the birds survived the extinction of the dinosaurs (June 6, 2014).
Electric fish: AC or DC?
October 12, 2013
Did you ever wonder whether electric fish emit AC or DC -- alternating current or direct current?
Now you know.
The group of figures above, from a new article, shows traces of the electrical discharges from two fish. You can see that the upper trace is AC, and the lower one is DC.
What makes this especially interesting is that these are two newly discovered species of fish, and they are closely related. They are considered sister species, within the same genus.
Some details...
The electrical traces show voltage (y-axis) vs time (x-axis). The x-axis scale bars are 1 millisecond. No scale is shown for the voltage (but it is around 1 volt). (Oddly, the y-axis is not labeled at all in the paper; the information I have given is from the news stories. How did a poorly-labeled graph such as this get published?)
These are discharges (bursts), not continuous currents. Nevertheless, the difference is clear, and it is something like the difference between AC and DC continuous currents.
The fish are two species of bluntnose knifefish from the Amazon: Brachyhypopomus walteri (top) and Brachyhypopomus bennetti (bottom). The scale bars for the fish are 1 centimeter.
As noted, the figures above are all from the article. The electrical traces are from Figure 2. The other frames of that figure show results for other specimens of the two species; the agreement within each species is good. The fish shown above are the top frames of Figure 3 (top fish) and of Figure 6 (bottom fish). There are numerous fish pictures in the article, including close-ups.
|
In fact, it was already known that some electric fish produce AC and some DC electricity. However, it was generally thought that DC was a feature of those fish, such as the so-called electric eels, that produce large jolts of electricity, which are intended to kill. Fish that produce weak electric fields have generally been thought to produce AC. The low voltage AC fields may be useful for communication and navigation. Thus the real surprise of the new work is finding two fish, apparently closely related, both producing weak fields -- one AC and one DC. Clearly we have limited understanding about why fish produce these two types of electric fields.
News stories:
* Electric Fish Kin Wired Differently. (The Scientist, August 29, 2013.)
* AC or DC? Two Newly Described Electric Fish from the Amazon Are Wired Differently. (Science Daily, August 28, 2013.) This news story includes considerable speculation about the roles of the two types of electricity for the fish. Emphasize that this is speculation, not anything that has been shown. The ideas discussed will guide further work.
The article, which is freely available: Two new species and a new subgenus of toothed Brachyhypopomus electric knifefishes (Gymnotiformes, Hypopomidae) from the central Amazon and considerations pertaining to the evolution of a monophasic electric organ discharge. (J P Sullivan et al, ZooKeys 327:1, August 28, 2013.)
More about biological electric fields:
* Cuttlefish vs shark: the role of bioelectric crypsis (May 10, 2016).
* Bees and flowers: A 30-volt story (June 21, 2013).
* Previous post about fish: Why might it be good to put lights on fish nets? (September 9, 2013). Actually, it's more about turtles.
* Next: Using a pH meter to help you find dinner (July 8, 2014).
Thanks to Borislav for suggesting this topic.
Why and how some cockroaches avoid glucose
October 11, 2013
To start, watch the following movie: Ten cockroaches look for dinner -- #2. (15 seconds; no sound.)
As you can see, they all chose the plate labeled fructose, and avoided the one labeled glucose. To understand the significance, now watch the following movie: Ten cockroaches look for dinner -- #1. (15 seconds; no sound.)
The movie titles are mine. However, I have numbered them as they are given with the article.
Why do we care what cockroaches want to eat for dinner? Both the context of the problem and the underlying biology are interesting.
The context of the story here is a practical one. Cockroaches are a nuisance. To combat them, we have developed cockroach traps, which feed the roaches a poison. The bait? Sugar. Glucose. Cockroaches are attracted to the glucose, and eat the poison in the trap.
Over time, cockroaches have developed resistance to the traps. Interestingly, this trap-resistance is not simply resistance to the poison, but it is an aversion to the trap itself. The trap-resistant roaches avoid the trap. Why? Because they are repelled by glucose, rather than attracted to it. The two movies above show how cockroaches respond to glucose. Movie 2 (which I suggested you watch first) shows the roaches that avoid the trap; they avoid glucose. Movie 1 is a control, with wild type (normal) cockroaches.
A recent article explores the basis of the response by the trap-resistant cockroaches. The authors refer to them as glucose-averse.
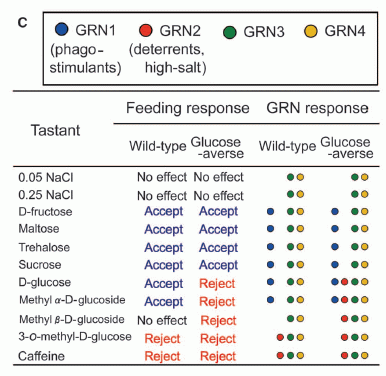
|
The table summarizes a lot of testing they did to figure out the underlying reason the mutant cockroaches avoid glucose.
The table includes two types of results for two types of cockroaches. The cockroaches are either the "wild-type" (normal) or the "glucose-averse" mutants. The two types of results are the "feeding response" and the "GRN response" -- the response of specific neurons.
Start with the feeding responses. You saw examples of the feeding responses for the two types of roaches in the movies above. The table here summarizes the results of many such experiments. You can see that the wild type roaches "accept" certain chemicals, including both glucose and fructose. They "reject" some others, including the notoriously bitter caffeine. The feeding responses for the glucose-averse cockroaches are similar -- except that they now reject a few chemicals that the wild type accepted; these include glucose, the focus of attention.
|
Now look at the GRN responses. What is GRN? GRN stands for gustatory receptor neurons; that is, GRNs are taste neurons. The scientists measure the electro-physiological responses of the insect equivalent of taste buds. Results are shown for four GRNs, but only two are of interest. (#3 and 4 are positive for all cases; they are controls.) For the wild type, you can see that GRN1 (blue dots) lights up for molecules that are accepted, whereas GRN2 (red dots) lights up for molecules that are rejected. The glucose-averse roaches show the same response -- except that there are three new red dots. Three new chemicals now show a response with GRN2 (red dots) -- the response that means aversion. These include glucose.
This is Figure 1C from the article.
|
In summary... Cockroaches can mutate so that they survive traps we set for them. Interestingly, one way they do this is by avoiding the glucose sugar that is added as the attractant. They avoid the glucose by an altered neurological response. For the glucose-averse cockroaches, glucose behaves as a bitter stimulant, and is therefore avoided.
News stories:
* Bittersweet: Bait-Averse Cockroaches Shudder at Sugar. (Science Daily, May 23, 2013.)
* Roach Motel Vacancies Explained -- Scientists discover why certain cockroaches avoid eating insecticide-containing sugary bait. (The Scientist, May 23, 2013.)
The article: Changes in Taste Neurons Support the Emergence of an Adaptive Behavior in Cockroaches. (A Wada-Katsumata et al, Science 340:972, May 24, 2013.)
More about cockroaches:
* Cockroach milk (August 21, 2016).
* Cockroach should be disinfected before eating it (February 12, 2013).
* Acrobatic cockroaches inspire robot design (September 16, 2012).
More about fructose and glucose: Fructose; soft drinks vs fruit juices (November 7, 2010).
More about flavor (taste + odor): How a cork causes an off-flavor in a beverage (October 21, 2013).
My page Organic/Biochemistry Internet resources has a section on Carbohydrates. It includes a list of related Musings posts.
October 9, 2013
Shale gas recovery using hydraulic fracturing (fracking)
October 7, 2013
In recent years a new tool for mining natural gas (and oil) has emerged. It's called hydraulic fracturing -- or, popularly, fracking. The idea is that water, with detergents and other chemicals, is pumped into the ground, with the goal of breaking up gas-bearing minerals. Fracking has opened up huge new supplies of natural gas. It has resulted in lower gas prices, and has affected geopolitics. And it has caused controversy. There are concerns about fracking. Some might be anticipated; some emerge with experience. Unfortunately, the public debate is often more heat than light. It's hard to figure out.
So what's with fracking? For some time, I have wanted to include an article on fracking, but never got around to it. I was preparing to write about one recent article -- and, while looking for a "news story" for it, came across a web page that is a quite good overview of fracking issues. This post is about that page; I'll skip the article.
The page's author, Richard Reeleder, considers five possible concerns about fracking. He examines the evidence, based on what has been published in the scientific literature. Let's look briefly at two of his points here.
One is the possibility that methane (CH4, the major component of natural gas) ends up in drinking water as a result of nearby fracking. The article I was going to include provides evidence for this. I'm not sure all the evidence is consistent with this, but that's ok for now. Interestingly, it seems that the appearance of methane in the drinking water is a sign of a bad well. Bad wells are a known problem; they should be brought up to code.
Another concern is that fracking might cause earthquakes. When discussing this, we need to make a distinction between two steps in the process: one is the fracking per se, and the other is the disposal of the waste water afterwards. It seems likely that the latter is more of a concern for quakes than the fracking itself. Is the problem significant? The jury is still out, perhaps, but it is likely that such problems are rare. Areas of fracking and water disposal need to be monitored. It is possible that some areas will be considered unsuitable for these activities.
Reeleder's analyses are short and rather well done. The five topics that he addresses are substantially independent; you can read one at a time. Read what he has to say with an open mind. I think the general picture that emerges is that we are all still learning. Problems should not be swept under the rug, but studied and managed, as with any large scale human operation. Unfortunately, the popular debate is too often dominated by those with a stake in one side, and presents extreme positions that are neither correct nor helpful. Reeleder is more interested in sorting out what we know, what we don't, and what we should do as a result. I hope you will browse his page.
Fracking: What the peer-reviewed science says about shale gas. (Richard Reeleder, NB datapoints, August 12, 2013.) The "About" page says that the site is written by a retired scientist. Importantly, it seems well written. My purpose is to put some complex issues on the table, not to promote an answer. That seems to be his goal, too. I think it serves that purpose well. It's better than my original plan, which was to focus on one article -- then noting that it is all controversial. (The article I had at hand is #11 on his list.)
More about hard-to-recover fuels: Converting Oklahoma to natural gas (October 1, 2008).
Also see:
* Fracking: the earthquake connection (June 19, 2015).
* Fracking: Implications for energy usage and for greenhouse gases (October 26, 2014).
* Quality of oil and gas wells -- fracking and conventional (August 18, 2014).
* Methane leaks -- relevance to use of natural gas as a fuel (April 7, 2014).
More about a possible connection between human activity and earthquakes: Groundwater depletion in the nearby valley may be why California's mountains are rising (June 20, 2014).
There is more about energy on my page Internet Resources for Organic and Biochemistry under Energy resources. It includes a list of some related Musings posts.
Underground hibernation in primates?
October 6, 2013
You may think of hibernation as a way to escape from the cold of winter. However, hibernation is more general than that: it's a way to escape from bad conditions. It's a good strategy so long as the bad conditions are expected to last for a while.
You may also think that hibernation does not occur with primates -- loosely, the monkeys and apes and humans.
A new article presents a new example of hibernation -- in lemurs. Lemurs are one of the oldest lines of primates, predating what we would call monkeys. It's not the first example of a hibernating lemur. What's really new here is that these lemurs hibernate underground.

|
A hibernating lemur. The ground cover on top of the lemur has been removed, to expose the animal.
This is a Sibree's dwarf lemur, Cheirogaleus sibreei, from Madagascar. The animal probably weighs about 250 grams.
(The fingers of a gloved hand are seen at the upper left.)
This is Figure 2 from the article.
|
The following figure is an example of what the scientists learn about the hibernation of these lemurs.
Temperature data during hibernation.
This is a rather complex figure, so let's go through it slowly.
The graphs plot temperatures (T, in °C, y-axis) vs time (x-axis), for two conditions. Each panel of the graph shows about a week of measurements; the light vertical bars mark midnight, and the short dark horizontal bars at the bottom mark darkness. (Dates are shown at the top of each frame; "19/5" means 19 May.)
The top frame is for a lemur in a "nest" in a tree; this is considered a poorly insulated place to hibernate. The bottom frame is for the same animal, now in a well-insulated place it has dug out below -- a hibernaculum. (Note that the dates for the two frames are consecutive.)
|
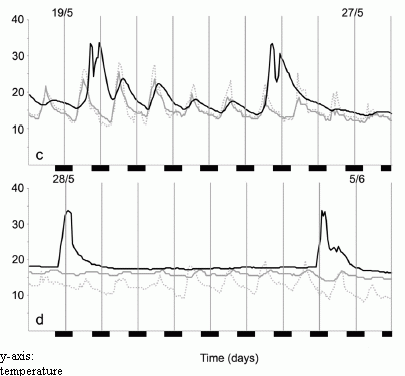
|
In each frame there are three sets of T data. The dark line is the skin temperature of the animal. The lighter solid line is the temperature where the animal is (tree nest or underground hibernaculum). The light dashed line is the ambient temperature.
In the top frame -- lemur in tree nest -- all three T curves vary with the daily cycle. Remember, this is a primate -- warm-blooded. Here its body T is tracking the external T; this is a hibernation state. (There are also a couple of times when the animal is aroused, and has a high body T. We can ignore those times -- though they are a reminder that most of the time the animal is hibernating.) It also seems that the ambient T is decreasing towards the end of this time period; perhaps that is the signal to the animal to seek better shelter.
In the lower frame -- lemur in underground hibernaculum -- the animal's T is rather constant (except for those occasional periods of arousal). The lowest curve there is for the ambient T, which varies through the daily cycle. The middle curve is the soil T near the animal; it's a bit warmer and more constant. The animal benefits from the warmer and more consistent soil T, and maintains a constant T just somewhat warmer.
This is Figure 1 parts c and d from the article.
|
Bottom line... This is an example of a primate -- a lemur -- hibernating underground. Why does it hibernate? The coldest T in the graphs is around 10 °C. The likely reason for hibernating is the seasonal food shortage.
News story: Primate Hibernation More Common Than Previously Thought. (Science Daily, May 2, 2013.)
Movies. There are two short movie files for the article. You can get to them from the article web page (below). Each is under one minute; there is sound (but nothing useful). Movie 2 is perhaps the better one, as it not only shows the hibernating animal, but includes a good view of its face.
The article, which is freely available: Underground hibernation in a primate. (M B Blanco et al, Scientific Reports, 3:1768, May 2, 2013.) It's a short, readable article. The first page is a nice overview of hibernation issues.
More on hibernation and such...
* What if a hummingbird's body temperature dropped to below 10 °C on a cold night? (September 29, 2020).
* Bears hibernate -- but stay warm (April 30, 2011).
More on early primates, such as the lemurs... Tarsier; eukaryotic cells (August 31, 2009).
and ... Hybrid formation between organisms that diverged 60 million years ago (May 8, 2015).
More on dwarfs: Ceres is leaking (February 18, 2014).
More from Madagascar: New evidence on the human colonization of Madagascar (September 16, 2016).
Also see... Were dinosaurs cold-blooded or warm-blooded? (August 23, 2014).
Update December 15, 2023...
Primates went extinct in North America about 34 million years ago, when the climate suddenly cooled to make the area inhospitable for them. The discovery of lemur-like Ekgmowechashala in the more recent fossil record of North America presented a mystery. Was it possible that one group of primates had actually survived what was thought to be a continent-wide extinction? A new article. based on new fossil finds from North America and Asia, suggests that they came in a separate migration via Asia, crossing the Bering land bridge. They, too, went extinct in North America, about 25 million years ago -- leaving the continent without primates until the arrival of Homo sapiens, presumably also via Asia. (The name of these animals means "little cat man", in Sioux. One might wonder how a more accessible name would affect the attention they get.)
* News story: Fossils tell tale of last primate to inhabit North America before humans. (Phys.org (University of Kansas), November 6, 2023.)
* The article, which is open access: Phylogeny and paleobiogeography of the enigmatic North American primate Ekgmowechashala illuminated by new fossils from Nebraska (USA) and Guangxi Zhuang Autonomous Region (China). (Kathleen Rust et al, Journal of Human Evolution 185:103452, December 2023.)
The rotifers that lack sexual reproduction: what do we learn from the genome?
October 4, 2013
This is a follow-up to an earlier post on bdelloid rotifers [link at the end].
These are interesting little animals; they manage to thrive without sexual reproduction. Sexual reproduction serves to purify the genome of harmful mutations. It is commonly thought that loss of sexual reproduction in a complex organism sets it on a path to extinction. The rotifers challenge that view.
A lingering question is whether the rotifers might have some low level of sexual reproduction that no one has observed.
A new article reports the genome sequence of a bdelloid rotifer, Adineta vaga. It offers some clues about rotifer asexuality. It turns out that meiosis -- a key step for sexual reproduction -- cannot occur in this rotifer at all, as the genetic make-up doesn't allow it. Adineta vaga is tetraploid -- in that it seems to have four copies of each gene. However, the genes have been oddly scrambled along the chromosomes, so that there are no homologous chromosomes. Without the pairing of homologous chromosomes, meiosis is impossible. Without meiosis, sexual reproduction is impossible.
The following cartoon, from the new article, is a nice presentation of this issue. It presents an important idea in biology, and shows how the rotifers fail.
In sexual reproduction there is an alternation between haploid and diploid states. Look at the left side of the figure. You see "meiosis" (bottom arrow) as the transition from diploid to haploid, and "fertilization" (top arrow) as the transition from haploid to diploid. A key step is the alignment, or "pairing", of the two homologous chromosomes in the diploid, and then their separation to produce haploid gametes (eggs and sperm). Particularly note how the gene order (A-K) is the same in the two chromosomes in the diploid state; this alignment is the basis of chromosome pairing, and is indicated here by dotted lines between copies of the same gene.
The right side shows, in cartoon form, what has happened to the rotifer genes. You'll recognize some of the same genes. There are lots of dotted lines, connecting pairs of the same gene. But there are no homologous chromosomes -- the kind seen on the left side. That means there can be no meiosis.
This is Figure 5 from the article.
|
Bottom line... The genome of this rotifer is scrambled. That is inconsistent with it having any sexual reproduction.
News story: How to survive without sex: Rotifer genome reveals its strategies. (Phys.org, July 22, 2013.)
The article, which is freely available: Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. (J-F Flot et al, Nature 500:453, August 22, 2013.)
Background post about rotifers: Lesbian necrophiliacs (March 8, 2010). Includes music.
A post that deals with meiosis: Children with two fathers (January 3, 2011).
There is more about genomes on my page Biotechnology in the News (BITN) - DNA and the genome. It includes a list of Musings posts on sequencing and genomes.
Borislav suggested this item, and included a first draft that served as the basis for this post.
October 2, 2013
Artificial brain-like structures grown from human stem cells in the lab
October 1, 2013
The following figure, from a new article, is getting a lot of attention.
|
What is it? It's a piece of brain tissue. Human brain tissue.
It was grown in the lab from stem cells.
|
The scale bar (upper left) is 200 micrometers. That is, the entire piece is about 2 millimeters across.
This is part of Figure 1a from the article. The full figure outlines the procedure, and shows samples from other stages of the process.
|
The authors call these things cerebral organoids.
What they did is straightforward, though of course technically complex. They started with stem cells -- induced pluripotent stem cells (iPSC). They induced the stem cells to become brain cells. They enhanced simple procedures to allow the tissue to organize. The result: small pieces of brain tissue -- grown in the lab from isolated stem cells.
In one sense, this is not surprising: making organized tissues from stem cells is a major emerging field of biology these days. However, this is novel: brain tissue. That gets our attention.
We'll leave the details of how they did it and how they characterized the complex brain tissue that developed. Instead, we'll note an example of how they can use this system, even in this early stage of development, for research.
In this experiment, they start with cells from two types of people. One is normal, or "control". The other carries a mutation that results in a defect of brain development, called microcephaly. They get stem cells from both people; for the control, they actually use commercial iPSC. They then make cerebral organoids from both types of stem cells.
|
Here is what they found. The figures show what the developing cerebral organoids looked like at various times.
The top set of pictures is for the normal cells, labeled "Control". The bottom set is for the microcephalic cells; it's labeled "A3842" (a code number for the particular patient).
This is Figure 6d from the article.
|
The observation: they are different. The article includes more data, and the authors interpret some of the details in terms of the disease. For now, we just note that the two kinds of cells behave differently. That is, a mutation that affects brain development in vivo affects the development of the brain tissue in this lab procedure. Thus this experimental system can be used to study how the mutation affects development.
A final comment... The pieces of tissue stop growing at about the size shown above. Why? Because they lack blood supply, needed to feed the cells inside the tissue mass. This is a common problem with the development of tissues in the lab. People are working on this problem. What will happen to the cerebral organoids if the problem of supplying nutrients is solved? Stay tuned.
News stories:
* 3-D Model Brain Built With Stem Cells: Organoids' Architecture Mirrors The Real Thing. (Medical Daily, August 28, 2013.)
* Human Brain Tissue Grown in Test Tubes -- The technique used to develop mini brains offers potential for establishing model systems for brain disorders. (GEN, August 28, 2013.)
* Lab-grown model 3D brains -- "Cerebral organoids" can model complex human brain disorders and the earliest stages of brain development. (Kurzweil, August 30, 2013.)
* News story accompanying the article: Developmental neuroscience: Miniature human brains. (O Brüstle et al, Nature 501:319, September 19, 2013.)
* The article: Cerebral organoids model human brain development and microcephaly. (M A Lancaster et al, Nature 501:373, September 19, 2013.) There is a preprint freely available at: pdf, preprint.
Using patient-specific stem cells to study Alzheimer's disease (February 24, 2012). In this post, we discussed work that used differentiated stem cells to study a brain disease. In some ways, it is similar to what was done in the new work. However, the new work involves a substantial step forward: not just differentiated cells, but developing tissue.
More on brain organoids:
* Brain development: comparing human and chimpanzee lab-grown brain organoids (November 5, 2019).
* Autism in a dish? (September 4, 2015).
More organoids: An organoid for the gut: at last, a culture system for norovirus (October 30, 2016).
There is more on stem cells on my page Biotechnology in the News (BITN) - Cloning and stem cells. It includes a list of related Musings posts.
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia). It includes an extensive list of brain-related Musings posts.
On handedness in humans
September 30, 2013
Briefly noted...
Handedness is interesting. For most people, one hand is better than the other at many tasks, such as throwing and writing. Why do we have this handedness? How does it develop in the individual?
A new article is a reminder of how little we know. The article suggests that many genes are involved, some of them being also involved in other body asymmetries. I note the item simply because of the topic: this post is something of an update on an interesting problem. However, there is no clear message to answer either question posed above.
Both news stories listed below are good, and provide useful readable overviews of the field. Emphasize some of the ideas being considered rather than any answers.
News stories:
* Gene Responsible For Left-Right Asymmetry Defects In Body Could Determine Handedness Too. (Medical Daily, September 12, 2013.)
* Genes linked to being right-or left-handed identified. (University of Oxford, September 13, 2013.)
The article, which is freely available: Common Variants in Left/Right Asymmetry Genes and Pathways Are Associated with Relative Hand Skill. (W M Brandler et al, PLoS Genetics 9(9):e1003751, September 12, 2013.)
More about handedness...
* Handedness in kangaroos: significance? (July 31, 2015).
* The origins of baseball -- two million years ago? (August 18, 2013).
* Analysis of teeth confirms that Regourdou was right-handed (September 7, 2012).
* On being ambidextrous (January 24, 2010). Good background -- even though it is not about humans. (The new work questions whether handedness phenomena in other animals is related to that found in humans. As with other issues, that must remain open.)
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia). It includes an extensive list of brain-related Musings posts.
Can a coral adapt to a more acidic ocean?
September 29, 2013
Adding carbon dioxide to the ocean makes it more acidic; it lowers the pH. This will make calcium carbonate, CaCO3, more soluble. What are the implications for organisms that make skeletons of CaCO3? We have discussed this before; the experimental results so far show that various animals respond differently to the lower pH. [A link to one such post is at the end.]
A recent article offers a new approach to addressing the question. The key point of the article is to examine how one species of coral responds to pH in nature -- rather than in lab experiments. The scientists found a natural environment where there is a variation in pH. Corals grow naturally throughout the area -- at different pHs. The scientists asked how the corals respond to the natural range of pH. They did this by measuring the CaCO3 skeleton from corals isolated from various pHs.
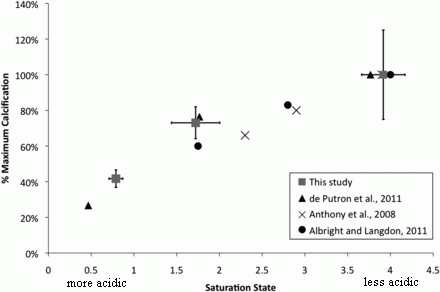
|
This figure summarizes the key findings. The graph plots the extent of calcification of the coral (y-axis) vs pH (x-axis).
The x-axis is actually more complex, but it represents pH -- with lower pH (more acidic) on the left. The x-axis scale was explained in the background post, but all you need here is the idea that higher acidity (due to more CO2) is at the left.
|
The graph combines results from multiple papers. Importantly, the points shown with squares (there are three of them) are from corals in the natural environment. These are the new results from the current article. The other points are from lab studies, published previously.
This is modified from Figure 3 from the article. I added the labels "more acidic" and "less acidic" along the x-axis.
|
The observation is simple: all the points seem to fit a single relationship. That is, the corals in the natural environment respond to increased CO2 and the resulting lower pH about the same as the corals in lab experiments.
This is an important result. However, it is also a limited result. It seems clear that the corals they studied here do not adapt any better than the corals in typical lab experiments. However, we do not know the limits of the conclusion. Are there features of this particular environment, perhaps the varying pH the animals are exposed to during their life, that make this a special case? Is it possible that the extent of skeleton formation is not very important to the coral, at least over the range studied here? What about other animals? The authors address some of the limitations. None of this diminishes the new article; it is a nice report. Over the long term we will find out how general the conclusion is.
News story: Underwater Springs Reveal How Coral Reefs Respond to Ocean Acidification. (Science Daily, June 17, 2013.)
The article, which is freely available: Reduced calcification and lack of acclimatization by coral colonies growing in areas of persistent natural acidification. (E D Crook et al, PNAS 110:11044, July 2, 2013.)
Background post: Increased CO2: effect on animals that make carbonate skeletons (January 11, 2010). This post has a graph with the same x-axis scale as shown above, and includes an explanation.
A similar analysis of another coral population: An example of coral growing well in a naturally acidified ocean environment (February 16, 2014).
More about coral skeletons: File dates and human settlement in Polynesia (November 16, 2012).
Also see:
* Ocean acidification and coral reef fishes: a controversy (March 16, 2020).
* When does global warming occur: day or night? (October 28, 2013).
Quiz: What are they?
September 27, 2013
I think we will continue with the new quiz format... A question will be posed, but the answer follows immediately, in the same post (rather than the following week). Those who want a week to think about the quiz will need to act accordingly.
|
The questions are about the figure at the left...
1. What are they? (That is, this is a pair of what?)
2. What animal is this from?
The figure is from the Cambridge University news story listed below. (It is similar to Figure 1D of the article.)
Want to see some action? Movie S2. It's about 3 MB, and runs 12 seconds. The action is slowed down -- about 150-fold.
|
Answers and discussion...
The figure shows a pair of gears. Perhaps you recognized that.
What might be more of a surprise is that these gears are from the legs of an insect, a leafhopper (or planthopper) called Issus coleoptratus. The gears are found in the larval stage.
These are functional gears, as you can tell from the movie, which is associated with a new article. The individual gear teeth are about 20 micrometers long.
What are the gears for? They serve to synchronize the rear legs during a jump. The synchrony achieved by using the gears is better than if the animal sent a nerve signal to each leg. They are, so far as the authors know, the first functional gears found in a living organism.
The article makes use of high-speed photography. The movie noted above was recorded at 5000 frames per second -- and played back at 30 frames per second, so you can see what is happening.
This is an incomplete story. The adult form of these insects do not have gears, yet jump as well or better than the larval stage. They seem to have other ways to achieve synchrony.
News stories:
* Got gears? Let's leap, says the leafhopper! (Why Files, September 17, 2013. Now archived.)
* From the host university: Functioning 'mechanical gears' seen in nature for the first time. (University of Cambridge, September 12, 2013.) It includes a promotional video, with brief science content and much about some community activities of the scientists. It is narrated by author Burrows.
The article: Interacting Gears Synchronize Propulsive Leg Movements in a Jumping Insect. (M Burrows & G Sutton, Science 341:1254, September 13, 2013.) The article has three movie files as Supplementary Materials; one was noted above. The movies should be accessible whether or not you can access the article.
More about gears:
* The Antikythera device: a 2000-year-old computer (August 31, 2011).
* Spinning gears -- driven by bacteria (February 1, 2010).
More about jumping insects: Jumping -- flea-style (February 21, 2011). This work is from the same people as the current work.
* Previous quiz: A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
* Next: Quiz: What is it? (September 23, 2014).
Thanks to Greg for suggesting this item.
September 25, 2013
Quantum gravity: the musical version
September 25, 2013
Video: Bohemian gravity. (Video at YouTube, 9 minutes.) Don't be intimidated by the title.
News story: This physics grad student made a mind-blowing Bohemian Rhapsody cover. (io9 (now Gizmodo), September 17, 2013.) Includes the video -- and the lyrics, and a link to the author's thesis.
More about (ordinary) gravity: The potato we call home: a study of the earth's gravity (May 3, 2011).
Previous music post: Stanford Linear Accelerator recovers 18th century musical score (June 22, 2013).
There is more about music on my page Internet resources: Miscellaneous in the section Art & Music.
Thanks to Jessica and Eric for sending this.
Computer scientist thinks; psychologist moves finger
September 24, 2013
A pair of humans has achieved what two rats did a few months ago [link at the end]. This achievement, by two professors at the University of Washington (Seattle) deserves brief noting, even though we do not have a scientific article yet.
What did they do? It's in the title of the post. One, a computer scientist acting here as the donor, had a thought. The other, a psychology professor acting as recipient, moved his finger. The connection? Brain-to-brain. The psychologist acted -- involuntarily -- as his brain received a signal from the brain of the donor.
Video. There is a video showing what the scientists did in carrying out this brain-to-brain test. It's called "Direct Brain-to-Brain Communication in Humans: A Pilot Study". It's short (1.5 minutes) and useful, but not very clear. You may need to play it a few times to put the pieces together, and you may need to pause it to read the information. The video is included with both stories listed below. It is also available directly at: video at YouTube. (There is no meaningful narration, though there is a bit of sound.)
News story: First human brain-to-brain interface. (Kurzweil, August 28, 2013.) This includes some interesting comments about the ethics of this work. Some of the comments challenge what the scientists said. Interesting.
Press announcement from the University: Researcher controls colleague's motions in 1st human brain-to-brain interface. (University of Washington, August 27, 2013.)
An article has been published; it is freely available: A Direct Brain-to-Brain Interface in Humans. (R P N Rao et al, PLoS ONE 9:e111332, November 5, 2014.)
Background post: Can one rat know what another rat is thinking? (April 8, 2013).
More from Seattle: The effect of delaying school start time on students: some actual data (March 12, 2019).
More about brains is on my page Biotechnology in the News (BITN) -- Other topics under Brain (autism, schizophrenia).
Where is the hottest part of a living cell?
September 23, 2013
You all know how to measure temperature (T). You put a thermometer in the object, and read the scale. You've probably seen various types of thermometers; the general idea is about the same, even if the technology within the thermometer varies.
What if you want to measure the T inside a living cell? Perhaps inside a mitochondrion? Perhaps you want to compare the T at various sites within a cell. And you want T with high precision, say to a hundredth of a degree.
A new article reports progress toward developing such a thermometer -- small enough and precise enough to be useful at the micrometer scale. It's a technology problem.
But first, some perspective. The following figure provides an overview of the capabilities of small thermometers.
The graph shows many types of temperature-measuring techniques, arranged here by their size (x-axis) and accuracy (y-axis). The high end of the size scale here is 10 µm, about the size of a typical animal cell. Methods suitable for use with living materials are shown in red.
One of the best of those "red" methods is labeled Nanodiamond. That is the method reported in a new article. The figure also shows a "Projected nanodiamond" -- a guess as to how good the authors think their method might be after further development.
This is Figure 1c from the article.
|
|
What did they do? The following cartoon figure is a useful diagram of the process.
|
We have a cell.
Start on the left, with the hexagon-shaped particle, labeled nanodiamond. This is a tiny diamond -- but not just any diamond. It's one known to have a special chemical impurity, a nitrogen vacancy.
The nanodiamond is irradiated with green light, shown coming from the top. Red light is emitted, by fluorescence; it is shown towards the bottom. What is important is that the exact color -- the wavelength -- of the emitted red light depends on the T, specifically the T of the nitrogen vacancy centers in the diamond. That's the basis of the thermometer; measure the color of the emitted light; it tells you the T. (We'll ignore the role of the microwave irradiation.)
|
The cartoon also shows how they tested this. There are actually two of the nanodiamond thermometers in this cell; the other is a little to the right. And very near that other nanodiamond is a piece of gold. The scientists shine a laser onto the gold, to heat it. That is, the gold particle is a hot spot in the cell; they have made it a hot spot by an artificial manipulation. They then measure the T in the cell at two spots -- at those two nanodiamonds. The one near the gold -- near the heat -- gives the higher reading. You can see that, in cartoon style, by the level in the little thermometer symbol next to each nanodiamond. That's the expected result. The nanodiamond thermometer works.
This is Figure 1 from the Sokolov news story accompanying the article.
|
The article contains real results from real cells, as hinted above. The presentation of the results is not easy to follow. If you can follow the idea in the diagram above, that is good.
What's the take home lesson from all this? It's certainly razzle-dazzle technology. They do seem to have achieved their goal, at least in part, of measuring temperature differences inside a small and delicate living cell. They discuss some limitations and, as noted with the top figure, are optimistic about future developments. They have opened the door to some measurements we might not have thought practical. Time will tell whether they can make their system practical.
News stories:
* Researchers develop nanodiamond thermometer to take temperature of individual cells. (Phys.org, August 1, 2013.)
* New Diamond and Gold-based Techniques Let Scientists Measure and Control the Temperature Inside Living Cells. (DARPA, August 5, 2013; now archived.) From one of the project sponsors.
* News story accompanying the article: Nanotechnology: Tiny thermometers used in living cells. (K Sokolov, Nature 500:36, August 1, 2013.) Check Google Scholar for a freely available copy.
* The article: Nanometre-scale thermometry in a living cell. (G Kucsko et al, Nature 500:54, August 1, 2013.) Check Google Scholar for a freely available copy.
More about thermometers:
* Heat production by aggregation of amyloid-beta in Alzheimer's disease (September 13, 2022).
* Hottest temperature ever recorded on Earth? Libya or Death Valley (California)? (June 30, 2013).
* Global warming trend? Independent evidence (March 22, 2013).
Another use of the fluorescence of nitrogen vacancies of diamonds: The smallest radio receiver (April 4, 2017).
More about examining the insides of a cell: The smallest tweezers -- for pulling single molecules out of cells (March 30, 2019).
Where is the MERS virus coming from?
September 22, 2013
We recently noted the emergence of a new viral disease, now called Middle East Respiratory Syndrome (MERS) [link at the end]. The virus is called MERS-CoV, with the CoV part of that denoting that this is a coronavirus. The virus is of concern because of its high fatality rate, but the number of cases is still low. Most cases so far have come from Saudi Arabia and nearby countries; most -- if not all -- of the others are easily connected with that area, either by direct travel or by likely contact with an infected person. There is evidence that the virus can be transmitted human-to-human; however, it is likely that most cases so far do not come from other humans. The source of the virus remains unknown.
A flurry of new reports suggests where the MERS virus may be coming from. Unfortunately, they don't really agree. Leading candidates include bats and camels. However, the evidence is limited.
The clearest finding comes from a survey of animals in an area around one of the cases from Saudi Arabia. The scientists report finding one specific sequence that matches the MERS virus in a sample from a local bat. Since other coronaviruses are associated with bats, this finding is thought to be significant. However, the evidence is very limited at this point -- a fragment of one gene and one bat.
The camel finding is even more limited. Antibodies to the MERS virus are found in local camels. That's about it. Are the camels carrying MERS-CoV? If they are carrying the human pathogen, are they involved in disease transmission, or are they merely bystanders? These are questions for further work.
This is clearly still a story in progress. Emphasize that the answer may be complex. One issue is where the natural "reservoir" of the virus is. However, transmission from that reservoir to humans may be indirect and complex. For example, it is plausible that bats might be the primary reservoir -- the major natural source of the virus. However, camels or other animals may be intermediaries in transmission to humans.
News story: MERS-CoV found in bat; hunt for other sources goes on. (CIDRAP, August 21, 2013.)
The bat article, which is freely available: Middle East Respiratory Syndrome Coronavirus in Bats, Saudi Arabia. (Z A Memish et al, Emerging Infectious Diseases 19:1819, November 2013.)
Another article -- a short and readable overview of the status of MERS. It is freely available: The Middle East Respiratory Syndrome -- How Worried Should We Be? (S Perlman, mBio 4(4):e00531-13, August 20, 2013.) It includes mention of the camel work.
It has been about one year since the first report of what we now call MERS. As of September 7, WHO reported 114 confirmed cases of MERS, with 54 deaths. The Middle East respiratory syndrome coronavirus (MERS-CoV) - update (now archived). A caution... There is no way to know the true total count of people infected. Only people who present to the medical care system are tested. We do not know if there is a vast reservoir of infected people with mild or no symptoms. This general problem is common with new infections, including for flu virus strains.
Background post about this virus: A new SARS-related virus seems to be emerging -- and an "ethics" story (February 4, 2013).
More MERS... Do camels transmit MERS to humans? (January 21, 2015).
Also see: Bats and the origin of SARS (January 25, 2014).
Two sections of my page Biotechnology in the News (BITN) -- Other topics are relevant here. One is on the general topic of Emerging diseases (general). One is on SARS, MERS (coronaviruses). SARS caused a human disease outbreak in 2003; the section has since been expanded to include MERS.
Previous post about bats: On a similarity of bats and dolphins (September 15, 2013).
Previous post about camels: Cloning: camel -- update (June 11, 2012).
Previous post about Saudi Arabia (other than the background post for this virus, listed above): The oldest known plants (November 2, 2010).
Brown fat: different kinds respond differently to cold
September 20, 2013
In a 2009 post, we introduced the idea of brown fat, with some recent developments [link at the end]. Brown fat tissue burns fat, producing heat but no metabolic energy. In contrast, common white fat tissue is for storing fat; the stored fat can be burnt for metabolic energy. An intriguing aspect of brown fat is that it might be good for weight loss -- an idea that has been validated in mice, but not in humans. The recent developments noted there included the finding of brown fat in adult humans, and the realization that brown fat in mice was more related to muscle than to white fat.
It now seems that the story is more complex. I should stress that nothing in the new work overthrows the basic findings reported earlier; it just provides a more complete picture of what is going on. It now seems that there are three types of fat, with two types of "brown" fat. One type of brown fat is the muscle-related brown fat noted above. But there is also some brown fat in white fat; this has led to the term beige fat. It seems likely that this type of brown fat is derived from the white fat -- not from muscle-related cells. The new work here shows interesting physiological differences between the "traditional" brown fat and the brown-white (beige) fat. Both can burn fat, and both can be stimulated by cold; however, they are regulated differently. Understanding brown fat, with the possibility that we might manipulate it for our benefit, requires understanding the two types of brown fat.
A new article explores the differences between the two types of brown fat. Here is one experiment from this new article.
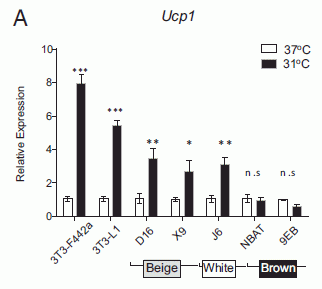
|
This experiment is done with cell lines, in lab cultures -- not in animals. The various cell lines represent the various types of fat cells; see the labeling at the bottom along the x-axis. (We'll ignore the ones that aren't labeled by the type of fat.)
For each cell line, there are two conditions, warm (37 °C; open bars) and cold (31 °C; solid bars).
The y-axis shows the level of expression of a key gene involved in the thermogenic (heat-generating) response. The scale is "relative"; for each cell line, the result at 37 °C is set equal to 1. That is, all the open bars are set to 1.
|
What do we see? Look at the height of the solid bars. For the brown fat, they are about 1 -- about the same as the open bars (or even less). For brown fat, cold does not seem to induce thermogenesis. However, for the beige fat and the white fat, the solid bar is much higher than the open bar. For these fat cell types, cold does induce thermogenesis.
This is Figure 3A from the article.
|
An important point is that this work is with cell cultures. There is no nervous system here. The response to cold is direct. The white and beige fat are directly stimulated by cold. Earlier work -- with animals -- had shown that brown fat was stimulated by cold; the nervous system was involved.
The conclusion? The key conclusion is that the two types of brown fat are regulated differently.
Significance? That's not clear at this point. The new work leads to a better understanding of the various types of fat, and how they respond to cold. The differences are interesting. All the facts need to be taken into account if we are to learn how to use brown fat for weight control. Can we exploit the differences between the types of brown fat? We'll see. At least we now know about the differences. Remember, only a few years ago we did not realize that adult humans had brown fat; now we know they have two types.
News stories:
* Fat Cells Feel the Cold, Burn Calories for Heat. (Science Now, July 1, 2013.)
* Temperature-Sensing Fat Cells. (The Scientist, July 1, 2013.)
The article: Fat cells directly sense temperature to activate thermogenesis. (L Ye et al, PNAS 110: 12480, July 23, 2013.)
Background post: Brown fat (May 21, 2009).
More about brown fat:
* An obesity gene: control of brown fat (October 2, 2015).
* Why exercise is good for you, BAIBA (March 10, 2014).
For more about lipids, see the section of my page Organic/Biochemistry Internet resources on Lipids.
September 18, 2013
More genome sequencing for newborns?
September 17, 2013
Briefly noted...
A few years ago we noted a case where genome sequencing for a child resolved a medical mystery, and seems to have led to proper treatment. At the time, such genome sequencing was too expensive to be routine. Since then, the cost of genome sequencing has plummeted. [Links to background posts are at the end.]
We now have an announcement from the US National Institutes of Health (NIH)... They are funding projects to study what the role of genome sequencing of newborns should be. This is a "big deal". We are at least near the point where such screening could become routine. However, we do not know how to use it effectively. That's the point. The NIH announcement is about asking questions; the purpose of this post is to raise those questions for you.
The announcement: NIH program explores the use of genomic sequencing in newborn healthcare. (NIH, September 4, 2013.) As you look over this announcement, note the diversity of projects they are funding.
News story: Sequenced at Birth -- An NIH program will study the promises and pitfalls of sequencing newborns' genomes. (The Scientist, September 5, 2013.) This links to other news stories.
Background posts:
* Genome sequencing to diagnose child with mystery syndrome (April 5, 2010). Links to a major update.
* The $1000 genome: Are we there yet? (March 14, 2011).
There is an extensive list of Musings posts in the broad area of personalized medicine: Personalized medicine: Getting your genes checked (October 27, 2009).
Perspective... DNA sequencing: the future? (November 7, 2017).
There is more about genomes and sequencing on my page Biotechnology in the News (BITN) - DNA and the genome. It includes a section with an extensive list of relevant Musings posts.
Sandstorms and midair collisions
September 16, 2013
How sandstorms work is not well understood. A new article uses computer modeling to explore what goes on during a sandstorm. Do collisions between sand grains in the air serve to strengthen the storm or to weaken it? It's an interesting story -- right or not.
|
Laying the groundwork for a sandstorm...
In this figure, each ball represents a grain of sand. The colors distinguish grains with different roles, as discussed below. The lines show the trajectories of certain grains. The big gray arrow (at the right) shows the wind direction.
|
The block of blue balls is the ground. Look at the yellow sand grain (ball)... Its trajectory is complicated: it goes down, hits the ground, and bounces back into the air. This collision of the yellow sand grain with the sand on the ground is the key event for this figure. Not surprisingly, the yellow grain stirs up a lot of sand -- and trouble -- when it collides with the ground.
Among the products of that collision... creepers, shown in green, which stay at the surface; and leapers, shown in red, which rise into the air, and are blown by the wind.
This is reduced from the figure in the Phys.org news story. It is probably the same as Figure 1 from the article.
|
That's just the background. There are now lots of particles in the air. They may collide with others in the air. That's the part that particularly interests the authors of the new work. They make a computer simulation to show what happens when sand grains collide in midair. The answer? It depends. In particular, it depends on how "bouncy" the sand grains are. They find that particles with the bounciness of real sand grains tend to stay in the air. That is, they think that midair collisions of sand grains serve to keep the sand in the air -- thus enhancing the storm. If, in their model, they turned off midair collisions, it reduced the strength of the storm.
This main conclusion is contrary to common wisdom, which says that midair collisions cause loss of sand from the air. So who is right? We'll see. If their model is correct, midair collisions enhance the sandstorm. But maybe they don't have it all right here. We'll see how people respond. People will study the model. They may find a bug, or they find uncertainties. They may improve the model, or they may question some of the parameters used. Such further study may help to resolve how sand grains behave in midair collisions.
News stories:
* Researchers discover midair collisions enhance the strength of sandstorms. (Phys.org, August 9, 2013.)
* Tiny Collisions Power Sandstorms. (Physics Central, August 9, 2013. Now archived.)
The article: Midair Collisions Enhance Saltation. (M V Carneiro et al, Physical Review Letters 111:058001, August 2, 2013.) A preprint is freely available at arXiv: arXiv preprint.
More about the properties of sand:
* How to climb a pile of sand (November 7, 2014).
* Playing in the sand (March 26, 2011).
More about collisions: Baseball physics (July 31, 2011). This post discusses the role of the coefficient of restitution (COR) in determining what happens in a collision. Restitution is a property relating to how elastic, or "bouncy", the colliding objects are. It's important in the current paper, but the authors don't explain what it is.
On a similarity of bats and dolphins
September 15, 2013
Bats and cetaceans (whales and dolphins) are both mammals. Unusual mammals, perhaps: one flies, one swims. Interestingly, some members of each group share an uncommon trait: they use echolocation. They send out a signal, and measure how long it takes for the echo to return. It's like radar.
A new article reports genome analysis of echolocating bats and dolphins -- and finds that both groups have similar genetic changes that allow the rather unusual sensory ability. That's an intriguing finding.
The work got started somewhat accidentally. Scientists noticed that one particular gene involved in hearing -- and therefore presumably in echolocation -- was surprisingly similar in a bat and a dolphin. Bats and dolphins are not very closely related (beyond both being mammals). Finding a particular gene where they shared similarities was striking. Did they share this gene similarity because it was involved in echolocation? And because these two organisms did echolocation about the same way? If they shared similarities in one gene involved in echolocation, what else might they share?
It was that last question that led to the genome analysis in the new article. What more similarities might be present? To address this, the scientists compared all available bat and cetacean genomes; some were determined specially for this work. They looked at members of each group that did and did not echolocate. As typical of modern genome work, computers analyzed the data. What the scientists found was that echolocating bats and echolocating cetaceans had about 200 genes with surprising similarity. In some cases, they knew the function of these genes; in other cases, the function is currently unknown.
That last point is important. Finding genes that are similar between the diverse echolocating mammals might suggest that these genes are involved in echolocation. Studying these genes may be a good way to learn more about the process. Or to learn about other -- previously unknown -- similarities between bats and dolphins.
Echolocation in diverse mammals is an example of convergent evolution: the same trait has arisen more than once, independently. That birds and bats both have wings is an example of convergent evolution; they have evolved the same function -- but obviously in different ways. The new evidence suggests that with echolocation, different mammals not only evolved the same function, but did it in about the same way. Interesting.
News story: Bats and Dolphins Evolved Echolocation in Same Way. (Science Now, September 4, 2013.)
The article, which is freely available: Genome-wide signatures of convergent evolution in echolocating mammals. (J Parker et al, Nature 502:228, October 10, 2013.)
More about echolocation:
* The use of wing clicks in a simple form of echolocation in bats (May 22, 2015).
* A plant that communicates with bats (September 7, 2011).
* What's around the corner? (January 7, 2011).
* Water: a bat's view (December 3, 2010).
More about cetaceans:
* Do dolphins talk to each other? (November 19, 2016).
* Effect of simulated sonar on whale behavior (March 16, 2014).
* A quasi-quiz: The fate of bone and wood on the Antarctic seafloor -- and the discovery of new bone-eating worms (August 20, 2013).
* Dolphins, bulls, and gyroscopes (September 10, 2010).
More about bats:
* Bats and the origin of SARS (January 25, 2014).
* Where is the MERS virus coming from? (September 22, 2013).
There is more about genomes on my page Biotechnology in the News (BITN) - DNA and the genome. It includes a list of Musings posts on sequencing and genomes.
The case of the missing incisors: what does it mean?
September 13, 2013
What do we learn from the following set of teeth?
|
This is a fossil human jaw -- an upper jaw.
This is Figure 1 from the article.
|
The key is to look at the missing teeth. However, that's more complex than you might think.
Start with tooth 11 -- an incisor. Next to it, labeled 13, is a hole -- a tooth socket; it's a socket for a canine tooth. The tooth itself is missing, but the scientists know quite well where the tooth goes, and what kind it is. Empty sockets such as #13 are not what really interests us. It's not surprising that an old fossil jaw has lost some teeth.
The kind of missing tooth that interests us is tooth 12 (labeled in red). Do you see why? If not, you might want to go look in a mirror. How many incisor teeth do you have? (Incisors are the teeth that look sort of like chisels; they are good for cutting. Tooth 11 is a good incisor. Next to the incisors are the canine teeth, with their sharp points. You should be able to count your upper incisors.)
Humans have four incisors. The subject of our study above, named B37, had only two. He was missing two incisors. It's not that two incisors are now missing from his fossil jaw, but that the jaw lacks places for two incisors -- teeth 12 and 22. Person B37 had a condition called maxillary lateral incisor agenesis -- or MLIA. That's a fancy term to say that the incisors are missing; agenesis means it wasn't generated. It's a known condition -- a genetic condition. B37 actually had bilateral MLIA, with the incisors missing on both sides.
Why is B37's bilateral MLIA of interest -- worthy of a published article? The scientists analyzed 28 fossil jaws from a particular site -- and found that 10 of them had this bilateral MLIA. That's more than one-third! They suggest that the high frequency of MLIA is evidence for substantial inbreeding -- endogamy -- in the community. What community? A farming community, in what is now Jordan -- over 9,000 years ago. This community dates back to the early days of farming. Is there some connection between the development of mankind's first farming communities and inbreeding?
One issue the authors address is whether the inbreeding might be simply due to being in a small isolated community. They argue that the archeological evidence suggests otherwise: the village was active in commerce. Thus they suggest that the inbreeding was a matter of choice, not necessity.
What should we make of this? It's interesting. There is a simple observation (missing teeth) -- and a simple interpretation (inbreeding). Then there are the bigger questions of interpretation. The situation is of historical interest. Are their facts correct? Let's hope so -- even though only a small number of jaws was examined. Are their interpretations -- at various levels -- correct? We have no way of knowing at this point; we would like to know what other evidence one way or the other can be found. If it's true, what is the significance? We'll leave that as a sociological question beyond our scope here. For now, we have this report, which is novel and interesting. What it means is open.
News story: First Farmers Were Also Inbred. (Science Now, June 19, 2013.) Includes good discussion of the interpretation of the findings.
The article, which is freely available: Earliest Evidence for Social Endogamy in the 9,000-Year-Old-Population of Basta, Jordan. (K W Alt et al, PLoS ONE 8(6):e65649, June 11, 2013.)
More about analysis of old teeth...
* The "hobbits": dentition suggests they were a distinct, dwarfed human species (November 30, 2015).
* How to eat if your jaw looks like a circular saw -- a follow-up (March 8, 2015).
* Bacteria on human teeth -- through the ages (March 24, 2013). Among other things, this post addresses the topic of early human farmers.
* Analysis of teeth confirms that Regourdou was right-handed (September 7, 2012). Neandertal teeth.
More from ancient Jordan: The oldest known bread (August 28, 2018).
More about farming:
* Microbes on your fresh fruits and vegetables? (May 29, 2013).
* Farming by amoebae (February 15, 2011).
September 11, 2013
False memories in the courtroom
September 10, 2013
A recent post was about false memories in mice [link at the end]. The topic is of general interest as a part of our understanding of the brain. However, the more specific issue of false memories in humans has been prominent in recent years, partly because of its relevance to our justice system. As an example, it has been shown that innocent people have been convicted based on testimony that was false -- not because the witness lied, but because the witness truly had false memories.
Nature recently ran a News feature on the issue of false memories in the courtroom. I encourage you to read this three page article. The goal is to raise awareness of the issue. Don't be surprised if you have complex feelings about the topic, or about the person they focus on.
News feature, which is freely available: Evidence-based justice: Corrupted memory -- Elizabeth Loftus has spent decades exposing flaws in eyewitness testimony. Her ideas are gaining fresh traction in the US legal system. (M Costandi, Nature 500:268, August 15, 2013.)
Background post: A mouse that remembers an event that did not happen (September 3, 2013).
More about the brain is on my page Biotechnology in the News (BITN) -- Other topics under Brain. It includes a list of relevant Musings posts.
More about justice: Police training (July 19, 2020).
Why might it be good to put lights on fish nets?
September 9, 2013
Large-scale commercial fishing is often done with large nets, called gillnets. Unfortunately, the nets also catch turtles. Scientists have considered the possibility of lighting the nets, taking advantage of the different visual systems of fish and turtles. A new article reports one such test, with encouraging -- but incomplete -- results.
The new work is done with ultraviolet (UV) lights. It is known that turtles can see UV, and many fish cannot. It is not known that this difference in vision would lead to the desired behavioral difference.
The following two figures summarize the key results from the article. Each figure compares the "catch" without vs with lights on the net. The lighting is provided by a "UV LED"; the non-lighted case is labeled "control".
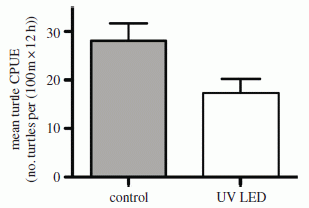
|
Turtle catch: catch-per-unit-effort (CPUE).
The "unit effort" is per 100 meters of net length and per 12 hours of capture time. This is the same normalization for both figures.
This is Figure 1 from the article.
|
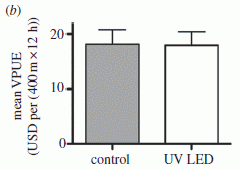
|
Fish catch, as judged by its market value: value-per-unit-effort (VPUE)
This is Figure 2b from the article.
|
The main observation is clear... The turtle catch (top figure) is lower with UV lights. The fish catch (bottom figure) is about the same regardless of lights. These are the desired results. Success, it would seem.
Indeed the result is encouraging, but we should note some limitations. First, the two results above are from different tests. They have not measured both responses in a single test. The lack of the internal control is a weakness of the experimental design. (There may have been a reason for them doing it first in separate tests. My point is not a criticism of what they did, but a limitation of the interpretation.) Beyond that, we do not know how general the result is. How reproducible is it? How well would it work in other regions of the ocean -- with different fish and turtles? And what is the optimum lighting? These additional questions are part of how new processes are developed. The current result is one test. It's encouraging, and further work is warranted.
News stories:
* UV lights reduce turtle bycatch. (Conservation Magazine, July 24, 2013. Now archived.) Brief.
* UV-Emitting Diodes Help Keep Endangered Sea Turtles Out Of Gillnets. (Science 2.0, August 6, 2013. Now archived.) Excellent overview.
The article, which is freely available: Developing ultraviolet illumination of gillnets as a method to reduce sea turtle bycatch. (J Wang et al, Biology Letters 9:20130383, October 23, 2013.)
For more about turtles:
* Magnetic turtles (July 5, 2015).
* Where is turtle #92587? (February 22, 2011).
More about UV vision:
* Carnivorous plants: A blue glow (March 16, 2013).
* Butterflies and UV vision (June 29, 2010).
More on fishing nets: Cultural transmission of fishing techniques among dolphins (September 13, 2011).
More about LED lighting:
* UV LEDs for the inactivation of viruses? (January 11, 2021).
* Effect of artificial lighting on the environment (September 3, 2015).
* CFL and LED lights: energy-efficient, but toxic (March 3, 2013).
More fish: Electric fish: AC or DC? (October 12, 2013).
There is a section of my page Internet resources: Biology - Miscellaneous on Medicine: color vision and color blindness.
My page of Introductory Chemistry Internet resources includes a section on Lighting: halogen lamps, etc.
CRISPR: What's it doing to help bacteria carry out infections?
September 8, 2013
CRISPR. It's becoming quite a story, one of the more fascinating stories in science in recent years. The CRISPR story started with the discovery of a defense system protecting bacteria from virus infections. A key feature of CRISPR is that it learns; it's something like an adaptive immune system -- and that was a surprise in bacteria.
Musings has noted a couple of recent off-shoots of the basic CRISPR story. One involved finding a CRISPR system in a bacterial virus. One involved finding molecular biologists who have learned to exploit the CRISPR system for their own needs as a tool for genetic engineering. [Links at the end.]
A new article reports another role for CRISPR in bacteria: some bacteria need CRISPR in order to infect their animal host (such as mice). That is, the earlier work showed that CRISPR was a defensive weapon, protecting the bacteria against invaders. The new work shows that CRISPR is also an offensive weapon, helping the bacteria invade its host.
The two effects are due to the same basic mechanism. In the "usual" defensive role, the CRISPR system makes an RNA that targets a gene in the virus it is defending against. In the "new" offensive role, it makes an RNA that targets a bacterial gene -- a gene that would alert the animal host to the invasion, and thus prevent bacterial infection. That is, the bacteria, via CRISPR, make an agent to counteract the host defense.
The new work is a study of Francisella novicida bacteria, which infect mice. Other Francisella are important human pathogens.
|
This graph illustrates the importance of the bacterial CRISPR system for causing disease in mice. It shows survival of mice (y-axis) over time (x-axis), after infection with various strains of the bacteria.
There are several data sets, but they really fall into two types. In one type, the mice are dead within 5 days; in the other, the mice survive fine.
|
First, look at the curve for wild type bacteria (WT; black line). It's one of those where the mice die. Next, look at the curve with the solid red square, labeled "Δcas9"; these bacteria are deleted for Cas9, a central player in the CRISPR system. With Cas9 deleted and CRISPR inactive, the mice survive the infection. Finally, look at the curve with the open red square, labeled "cas9 complement". With Cas9 restored, the mice die.
This is Figure 4b from the article.
|
That is... Normal bacteria kill the mice. Remove Cas9 -- and thus remove CRISPR, and the bacteria do not kill. Add back the Cas9, and the bacteria kill. That's good evidence that Cas9 is needed for the killing. (The reason for doing the "Cas9 complement" test, adding back Cas9, is that it helps makes sure that the previous test was doing what we thought. Imagine that something had "gone wrong" in the construction of the Δcas9 strain, and that it wasn't what we thought. Adding back cas9, which should reverse the effect, is a control.)
The other parts of the figure test more parts of the system. We'll skip the details here.
The scientists are able to show that what the Cas9 is doing is limiting production of a bacterial product that the animal host recognizes as a sign of infection. That bacterial product (a lipoprotein) stimulates the host immune system. Cas9 limits production of the bacterial product -- and thus limits the host immune response. That promotes infection.
So we see that CRISPR plays both offense and defense. Above we referred to one as its old role and one as new. That refers to our order of discovering them. Which came first in nature? Who knows. In terms of how they work, they are the same basic system -- just targeted differently.
In both cases, the CRISPR system acts within the bacterial cell. The bacteria make an RNA molecule that targets the gene to be inactivated. In one case, that target is a phage trying to infect the bacteria. In the new case, the target is an endogenous bacterial gene; reducing the expression of that bacterial gene promotes infection of the animal host, by counteracting the host defense.
News story: Bacterial security agents go rogue. (Emory News Center, April 15, 2013.) From the University where the work was done.
The article: A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. (T R Sampson et al, Nature 497:254, May 9, 2013.)
Background posts about CRISPR:
* Exploiting the bacterial immune system as a tool for genetic engineering: The Caribou approach (May 4, 2013).
* A virus with an immune system -- stolen from a host? (March 25, 2013).
More... CRISPR: an overview (February 15, 2015).
Other examples of complementation:
* How rice recognizes a Xoo infection (August 28, 2015).
* Do human genes function in yeast? Yeast-human hybrids. (August 21, 2015).
Upsalite: a novel porous material
September 6, 2013
Porous materials are of interest. They may be useful for adsorbing things, such as moisture.
A new article reports a new porous material. The properties of the new material seem promising, and there is an amusing story behind making it.
|
The figure shows the ability of this new material to adsorb moisture, and compares it with other materials.
The graph shows the amount of water adsorbed (y-axis) vs the relative humidity (RH; x-axis). Results are shown for four materials.
The measurements are made by first increasing the RH, then decreasing it. Thus the curve for each material is double; arrows show the direction of RH change. In each case, water adsorbed increases with increasing RH, then decreases as the RH is reduced. However, the amount lost as the RH is decreased is less than what it gained on the "way up". That is, the amount of water held depends on whether RH is increasing or decreasing. We will come back to this in a moment.
|
Two materials (dark blue and black -- poor choice of colors!) show generally good adsorption of water. One of these is the new material, called Upsalite (blue). The other is a commercial material, called Zeolite-Y (black). In contrast, the other two materials tested here (red and green curves) show little moisture adsorption except at high RH.
Look more closely at the two upper curves, for Upsalite and Zeolite-Y. Both materials bind a lot of water; the Zeolite is a bit better at the high end. But now look at those arrows -- and go check that paragraph in fine print above. As we reduce the RH, the Zeolite loses most of the water it bound. In contrast, the Upsalite retains most of the water it bound at high RH as the RH is reduced. This is a useful property for a material intended to remove water. (The water is easily removed from the Upsalite by gentle heating.)
This is Figure 3C from the article.
|
What is this stuff, and what is the story of making it? The material is a form of magnesium carbonate, MgCO3. In particular, it is anhydrous and amorphous. "Anhydrous" means that it lacks water -- clearly a good idea if you want a material that adsorbs water. "Amorphous" means that it lacks regular structure; instead, its irregular structure has lots of holes (pores) in it, where something can be adsorbed.
The general approach for making it seems straightforward -- in a sense. The scientists suspend the readily available magnesium oxide, MgO, in methanol (methyl alcohol, CH3OH). They add carbon dioxide. A complex gel forms. They heat the gel to drive off the methanol; what is left is the desired product. The pores form where the methanol had been.
The problem with doing this is that it is well known that it doesn't work. Scientists tried doing it over a hundred years ago, and many times since.
It does work, according to the new article. How did they succeed where others had failed? Apparently, according to the news stories, they left the initial mixture sit over the weekend -- by accident. By Monday, the gel had formed. The procedure in the article specifies that the initial reaction should be given four days.
Thus, the story of discovering Upsalite is, in part, a story of an accident. Of course, they were doing careful thorough studies to see if they could solve the problem; the accident contributed one clue. The new material is promising. As the graph above shows, Upsalite is better at holding water, especially as the humidity is decreased, than the commonly used Zeolite. The scientists think that making Upsalite will be cheaper than making the more complex Zeolite; time will tell whether this holds up. We may not need to wait long; they have formed a company to develop Upsalite as a commercial product.
News stories:
* Upsalite: Scientists make 'impossible material'... by accident. (Phys.org, August 13, 2013.)
* 'Impossible' material made by Uppsala University researchers -- Ultra-adsorbing Upsalite material has the highest surface area for an alkali earth metal carbonate. (Kurzweil, July 19, 2013.)
The article, which is freely available: A Template-Free, Ultra-Adsorbing, High Surface Area Carbonate Nanostructure. (J Forsgren et al, PLoS ONE 8(7):e68486, July 17, 2013.)
Upsalite? It's named after the famous Uppsala University, in Uppsala, Sweden. We might also note that the work was done there in the Ångström Laboratory -- named after two of the university's former faculty, one of whom developed the unit of length that now bears his name.
More about materials to adsorb moisture:
* Harvesting water from "dry" air (July 1, 2017).
* What if your house could sweat when it got hot? (November 30, 2012).
More about porous materials:
* Cooperation: a key to separating gases? (March 28, 2014).
* Stanford scientists discover that ink sticks to paper (May 29, 2010).
A recent post about a carbonate: Photosynthesis that gave off manganese dioxide? (July 21, 2013).
Another post about zeolites: Upgrading ethanol? (April 11, 2016). Zeolite is a term for a broad group of chemicals; there is no connection between the functions in the two zeolite posts.
and... A better catalytic sponge: degrading plastics, and more (August 11, 2020).
Thanks to Greg for suggesting this item.
A mouse that remembers an event that did not happen
September 3, 2013
It is a common type of study with mice and other animals... Scientists train an animal to learn a behavior, and then test it to see how well it has learned. In a new article, scientists instill into a mouse a memory of an event that did not happen. Subsequent testing shows that the mouse acts based on what it thinks it learned -- even though what it learned never happened.
It's an intriguing study, and the result is perhaps unsettling. It's also a difficult article. Our goal here is to try to give an idea of what the scientists did. Let's resist judging the significance.
Here is a key experiment...
|
In each test, a mouse was allowed to roam in a device with multiple chambers. The path the mouse explored was recorded; from this, the scientists can calculate how long the mouse spent in the left and right chambers.
The first test (top frame, labeled "mCherry") involved a control mouse. It explored both sides about equally. (The article reports quantitative data, but the visual impression from these mouse path records will serve our purposes here.)
In the second test (middle, "Label left"), the mouse feared the left chamber -- and therefore spent more time on the right. In the third test (bottom, "Label right"), the mouse feared the right chamber -- and therefore spent more time on the left.
This is Figure 4B from the article.
|
The results described above should make sense. (Honest. If they don't, try going through it again.) The key idea is that if a mouse fears one side, it avoids it -- and spends more time on the other side.
So what's the catch? If everything presented above makes sense, why is this of interest? If the mouse (say, the one in the middle) had had a bad experience in the left chamber, it would have a memory of the bad event, and would tend to avoid the left chamber, where the bad event happened. If. However, the mouse had not had any bad event in the left chamber. It did have a memory of such an event, but no such event had happened. That is, the mouse had a false memory -- and acted on the false memory.
How does a mouse get a false memory? The scientists used some "tricks" that have been developed over recent years. A caution... This is complicated, especially if the background steps are unfamiliar. The goal here is to give the idea, but it is not enough to fully explain what was done.
The key player is something called channel rhodopsin; note the "ChR2" in some parts of the figure. Channel rhodopsin is a protein that absorbs light. The scientists have modified brain cells so that they carry the gene for ChR2 -- and so that ChR2 is made when a memory is formed. When ChR2 absorbs light, it activates the neuron. That's some background. What did they do here? The following description closely follows that in the Kurzweil news story listed below.
Procedure:
Put the mice in a chamber; let's call it A. Let the mice become familiar with chamber A. As they do that, they form memories of chamber A. Because of the special construction of these mice, that means they make ChR2 in the cells used for forming those memories.
The next day, let the mice explore chamber B. While they are in chamber B, give them a mild shock -- and give them the light needed to stimulate the ChR2.
The next day, test the mice to see which chamber they fear (because of the shock). The answer is that the mice fear chamber A, not B. They fear chamber A because the light activated cells for memory of chamber A just at the time of the shock. The mice associate the shock with chamber A, because of the "trick" done by the scientists. They do not associate it with chamber B, which is where they got the shock. Clearly, the mice have a false memory; they associate the shock with chamber A, but actually were not shocked in chamber A.
This is what was shown in the figure above -- without any explanation of what was behind it. When we initially discussed the results above, we simply noted that the mice showed a preference for one chamber, based on a fear of the other chamber. We now see that the fear was based on a false memory.
What does this mean? In particular, does this have anything to do with false memories in humans? It's not at all clear what it means. Simply being able to do it is a technical achievement reflecting progress in understanding how memories work. The scientists have used novel experimental methods, and shown that they can control a mouse memory. However, it is premature to spend much time trying to interpret it. That's a common situation. Early work in a field is exciting simply because they can do it; it takes a while to work out the details and the understanding. This is something of a breakthrough experiment. Scientists will be studying it in detail, questioning each step of the procedure. And they will try to extend it and understand its significance.
News stories:
* Manipulating Mouse Memory -- Researchers deliver bursts of light to specific cells in the mouse hippocampus to implant false memories about a fearful event. (The Scientist, July 25, 2013.)
* Neuroscientists plant false memories in the brain -- MIT study also pinpoints where the brain stores memory traces, both false and authentic. (Kurzweil, July 26, 2013.)
The article: Creating a False Memory in the Hippocampus. (S Ramirez et al, Science 341:387, July 26, 2013.)
More about false memories: False memories in the courtroom (September 10, 2013).
Recent posts about memory:
* Long-term data storage in glass (August 14, 2013).
* Near-death experiences: are the memories real? (August 11, 2013).
More about the brain is on my page Biotechnology in the News (BITN) -- Other topics under Brain. It includes a list of relevant Musings posts.
Thanks to Borislav for suggesting this item.
Top of page
The main page for current items is Musings.
The first archive page is Musings Archive.
E-mail announcements -- information and sign-up: e-mail announcements. The list is being used only occasionally.
Contact information
Site home page
Last update: December 30, 2025