
| See the Chapter handouts and Homework section of the General Information handout (syllabus). | |
|
A. Introduction
B. Perspective: levels of organization C. Perspective: organism complexity D. Perspective: E. coli E. Perspective: chronology F. Mendelian genetics G. Genetic mapping H. Cells |
I. Chromosomes; cell division
J. When and where? K. Further reading L. Computer resources M. Errata N. Homework O. Partial answers Bottom of page; return links and contact information |
| Clark & Russell (see syllabus). (The 1st and 2nd editions of C&R seem to be organized essentially the same way, except for a couple of new chapters at the end of 2/e. If you happen to have the first edition, that is probably fine for all the basics.) I will briefly note relevant chapters at the start of the chapter handouts. However, since chapter correspondence between any two books is always incomplete, you will need to use some judgment. Please let me know of additions/corrections as well as usefulness of this information. Most important for our coverage here... Ch 3. Basic genetics. Also... Ch 1: general introduction to molecular biology. Ch 2: brief introduction to cells, mainly bacteria, and viruses. Fig 5.17 summarizes mitosis. Ch 11: first 2-3 pages is a useful introduction to eukaryotes. Ch 26: history, chronology. The Glossary may be useful at times, and also the Index. | |
Please read Weaver's Preface. He notes that he intends Ch 1-4 to be a review of what he considers to be background material. Our Extension students are quite heterogeneous. Each of you will need to deal with these introductory chapters in a way that is appropriate for your own background. However, one area that he assumes as background is genetics. My experience is that X107 students often have not had much genetics. (In fact, I am surprised that Genetics is a prerequisite for Molecular Biology, period.) Therefore, I will spend considerable time elaborating on the genetics part of Ch 1.
More generally, in discussing these introductory chapters, I will emphasize those topics that I think are most important background and which are not covered further. Some important topics are introduced in these chapters, then dealt with at length later. I will leave it to you to read the introductory material as appropriate for your own background, now and/or later.
We will not formally discuss Ch 4 at all, but can refer to it from time to time as needed.
Highlights:
Biology can be viewed at many levels of organization. Each level reflects the properties of lower levels, and their interactions.
Focus on cells as the basic unit of biological systems.
Looking "upward", cells organize into tissues, organs, organisms, populations, communities.
Looking "downward", cells are composed of organelles, macromolecules, small molecules, atoms.
In molecular biology, we are particularly concerned with the molecules: macromolecules and small molecules.
In molecular genetics we are particularly concerned with those molecules directly related to genetics: the molecules that are the genes, and that are involved with gene function.
Complexity of different organisms is expressed here as the amount of genetic material, in DNA base pairs (haploid genome). The table is in order by genome size. (Weaver has a similar table in Ch 2, p 34.)

The table shows that humans are about 1000 times more complex than E. coli, as judged by DNA content -- but only 7 times more complex as judged by gene number! The simple eukaryote, yeast (Saccharomyces cerevisiae), is only slightly more complex than E. coli.
Some plants -- and frogs -- have 10-100 times more DNA than we do!
It appears that increasing genome size (increasing complexity) may have two components. One is real complexity, the kind we would expect in going from bacteria to humans. The other is some kind of anomalous complexity, the kind we see in the amphibians, for example. (More about this "C value paradox" in Ch 2, p 36.)
At the bottom of the table are organisms that are simpler than E. coli. The mycoplasma are the simplest bacteria -- the simplest cellular, free-living organisms. That "tiny" Mycoplasma genome has stimulated discussion of the possibility of constructing an artificial cell (Hutchison et al, 1999). (E. coli is about 8-fold more complex than the simplest known cell.) Then some viruses. These are not free-living organisms, so their genome sizes do not represent the requirements for life. The gap from large virus to small cell is less than 3-fold in genome size.
The viroid genome could code for only 119 amino acids. That's barely one small protein. In fact, the viroid probably does not code for protein at all. We do know that the viroid is infectious.
The scrapie agent, called a prion, is even more mysterious. There is no evidence for any nucleic acid at all. See Schiermeier (2001), Fändrich et al (2001), Chien & Weissman (2001), and Saborio et al (2001).
Some of the genome sizes are exact, because these genomes have been entirely sequenced. Phage λ (lambda) has a genome of 48,513 base pairs -- and we know what they all are. The C. elegans sequence, marked with a *, is considered complete, although in fact it is not. A small portion of the genome, undoubtedly less than 1%, has proved difficult to sequence. This is probably gene-free DNA, mainly repetitive. (The issue of calling a sequence that is technically not complete as "complete" has provoked some controversy. This will increasingly be a problem with larger genomes. See Hopkin, 1999, for a discussion of what it means for a genome to be "complete".)
Weaver discusses the methodology of DNA sequencing in Ch 5, which is not on our core schedule for this course. Also see Ch 24, Genomics; this is new for the 2nd edition, and I have not read it yet.
Cell mass is about 1 picogram (10-12 g), or 6x1011 amu (daltons), 70% water.
About half of the dry weight is protein, about 2.4x106 molecules.
Genome size allows for 3600 (average-sized) proteins.
Actual genome sequence suggests 4288 genes.
About 2100 proteins have been displayed on gels (2-D).
Nearly 2000 genes have been identified by mutations.
About 800 small molecules have been identified.
Table 1.1 provides a sense of chronology.
The King & Stansfield Dictionary of Genetics includes an extensive chronology, from 1590 to 2005 (for 2006 edition). [This book is listed in the Syllabus, Supplementary books.] Also see Lander & Weinberg (2000).
(You are not responsible for history per se, such as names and dates.)
This is probably the most important section of the chapter for us at this time, along with Genetic mapping, Sect G, below. If you would like some supplementary materials, see the computer resources listed in Sect L.
"Transmission genetics". Characters behave as if they are "particles" (genes), 2 copies per individual (in higher organisms), randomly distributed to offspring: independent segregation. Mendel's First Law. (Weeks et al, 2001, present an unusual animal that is not diploid.)
Different characters are distributed independently: independent assortment. Mendel's Second Law.
Genes can exist in different states.
Terms: (Most are in the Glossary.)
Allele. A form of a gene.
Genotype, phenotype. The genotype is the information in the genes. The phenotype is the characteristic(s) we observe. The genes behave according to Mendel's laws.
Diploid, haploid. Diploid = 2 genome sets per cell. Haploid (or monoploid) = 1.
Homozygous, heterozygous. Refers to whether the two copies of a gene in a cell are the same or different. (The term hemizygous is sometimes used when there is only one copy.)
Dominant, recessive. If heterozygous, which allele determines the phenotype? If one does, it's called "dominant"; the other, "recessive", appears silent.
Mutant, mutation. A mutation is a change in the genetic information. A mutant is an organism carrying a mutation.
Wild type, markers. Used, loosely, for both genotype and phenotype. The wild type is supposedly the most common or normal allele (or characteristic), but the choice is sometimes arbitrary. [Weaver notes this, p 5, as he also offers the alternative term "standard type" -- which I think is less common.] Specific mutations (or their resulting mutant phenotypes) may be considered markers. The + symbol is often used to indicate the wild type -- or to indicate the presence of a property, which may or may not be the same thing. These are common and useful terms, but be careful that you know what they mean in each specific case.
Example. Consider methionine synthesis in bacteria. E. coli can make its own Met. Thus its
phenotype is Met+; this is the wild type. A mutant which can't make Met has
the phenotype Met-. The defect is due to a specific mutation, say metA98
or metB3, where the A and B indicate specific genes for methionine synthesis. A
diploid made from these two mutants may have the wild type Met+ phenotype,
even though it carries both mutations. Its genotype is
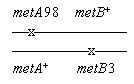
We would say that the two mutations complement (each other).
Exceptions
We do not always get the simple results that Mendel got. Examples of the complexities:
Dominance may not be clear-cut, or even constant.
Multiple genes may affect a particular character.
Genes that are on the same chromosome may or may not assort independently. They are linked, some are linked tightly. This is an important case, which we will discuss further; see Genetic mapping, Sect G, below.
And some more specialized exceptions...
The sex chromosomes represent a special case. Weaver discusses an example on p 3. (Hurst, 2001, discusses the sex chromosomes.)
Some genes on the chromosome don't have fixed positions. See Ch 23 for more about such "jumping genes" (transposons).
Some characteristics do not follow Mendel's laws at all. Cytoplasmic genes are in organelles such as the mitochondria or chloroplasts; these genes are (generally) maternally inherited. (See Awadalla et al, 1999, for some evidence that mitochondrial transmission may not be entirely maternal in humans.) Regardless, these genes are on DNA. Thus, at the molecular level, cytoplasmic genes are quite normal. At the cellular level they are different, because they are in an unusual location. See Wallace (1999) for a discussion of diseases due to mitochondrial genes, Michikawa et al (1999) for relevance to aging, Fliss et al (2000) for relevance to cancer diagnosis, and see Gray & Raybould (1998) for an interesting application.
We cross two parents with different genetic markers, then measure the frequency of the various kinds of offspring. The closer together two markers are on the same chromosome -- the tighter they are linked -- the more often they will appear together in the offspring.
Example... A+ B+ x A- B-
The cross will yield some parents, A+ B+ and A- B-, and some recombinants, A+ B- and A- B+.
If Mendel's 2nd law held universally, we would get 25% of each -- a total of 50% recombinants.
On the other hand, if A and B are very close on the same chromosome (i.e., "tightly linked"), we would get very near zero recombinants.
The frequency of recombinants is a measure of how close the two genes are.
Fig 1.4 illustrates such a cross. This, and the example above, are two-factor crosses.
Three-factor cross [not in book; important idea]
A three-factor cross involves three genes. What happens is a logical extension of what we have already seen.
In a three-factor cross, certain types of recombinants require two crossover events to occur; those are relatively rare. Example:
A+ B+ C+ x A- B- C-
We might find, for example, that A+ B+ C- and A- B+ C+ occur at a few percent, but that A+ B- C+ is very rare. This result would mean that the third class requires two events. The order shown in the example would fit that data.
The top half of Weaver's Fig 22.1 is useful. The top two frames diagram single and double crossover events.
Genetic mapping is logically very similar in complex and simple organisms -- in eukaryotes,
prokaryotes and viruses. You should be able to draw chromosomes aligned for recombination
and to determine the number of crossover events that must occur to achieve a particular
recombinant genotype. In particular, you should be able to predict the effects of single and
double crossover events.
Crosses with diploid organisms are more complicated to analyze, because there are two copies of each gene. That is, the genotype may be obscured (because you see only the phenotype).
The cell is the fundamental unit of life. Cells have the properties we associate with life: given food, including energy, they can reproduce.
The DNA molecule, in an information sense, can replicate, but not unless machinery is provided.
There are two major types of cells. Weaver discusses eukaryotic cells, e.g. p 4. Bacteria have a much simpler cell structure, called prokaryotic. I will show examples in class. Weaver only briefly notes the simplicity of prokaryotes (p 5). What prokaryotes lack is the broad spectrum of internal membranes, including the nuclear membrane. One can even say that bacteria do have a nucleus, though one that is simple compared to eukaryotic nuclei. The simplicity of bacterial cells is one reason so much work in classical molecular biology was done with them.
Martin & Muller (1998) propose a new model for how these two cell types are related. Gustafson et al (2000) may or may not be relevant, but is fun.
Schulz et al (1999) describe a new species of bacteria, with cells big enough to be seen by eye.
Beveridge (1999) reviews one distinctive aspect of bacterial structure.
Nurse (2000) reviews two centuries of cell biology.
We do not need much of the detail about eukaryotic cells that is presented. You should be generally aware of the complex intracellular membrane systems in eukaryotic cells. You should recognize the ER for its role in protein synthesis. You should be generally aware of the nuclear membrane and its pores. Refer to this material as needed later.
See Sect L for some Computer Resources about cells.
We will not discuss this section. You do need to be generally aware of chromosomes and cell division processes, but are not responsible for any details. We will discuss some aspects of chromosome structure in Ch 10-13.
The chromosomes are the physical carriers of Mendel's genes. There are two copies of each chromosome per (diploid) cell.
Mitosis (Box 1.2) is the basic process for distributing chromosomes at (eukaryotic) cell division. Prior to mitosis, the chromosomes duplicate; during mitosis, the two duplicates of each are sent off to the two daughter cells. Mitosis creates two daughter cells each genetically identical to the parent cell.
Meiosis (Box 1.3). The germ cells are haploid. Meiosis involves one round of chromosome replication and two cell divisions; the "extra" division reduces the ploidy. The original chromosomes replicate, producing four copies per cell. Then after two divisions, each meiotic product has one copy of each chromosome. And it is random which it has. This is the physical basis of Mendel's laws.
The two division steps in meiosis. The first division (anaphase I) separates the two homologs. Each chromosome at that point already has replicated (thus contains two chromatids). The two cells that result from the first meiotic division are not the same, because they get different homologous chromosomes. In the second division, the two chromatids separate, and are partitioned to the two daughter cells. Thus the two cells that result from each second division are identical. The second meiotic division is similar to a mitotic division.
Most analyses of meiotic products, as in ordinary genetic analysis, involve a random sampling from a large number of meioses. For a few organisms, such as yeast and Neurospora, one can analyze the four products of an individual meiosis.
Vale & Milligan (2000) explore how motor proteins, which move chromosomes, work. Willard (2000) discusses artificial chromosomes. Jallepalli et al (2001) discuss the fidelity of mitosis. Gachet et al (2001) discuss the control of mitosis.
We tend to focus on the what and how in molecular biology. But two additional issues are important and should be briefly noted here.
Many aspects of molecular biology are dynamic. Of course, we realize this over the large scale, as growth and division. But structures of individual molecules are dynamic, and so are the structures of assemblies, such as chromosomes, membranes or microtubules. These structures are held together by "weak bonds" (Ch 3), which individually are turned over rapidly. As a result, macromolecular structures vary in time due to random changes, and in response to interactions with other molecules.
The second special issue is location. Ultimately we need to deal with how the molecules of molecular biology get to their proper cellular location.
Some of the papers listed here are specifically referred to in other sections. Others are for general interest. I do assume that you read this "FR" section of the handouts; the annotations contain useful information. You have no responsibility to read any of the papers. (Also, remember that Weaver has his own "FR" update at his web site.)
For information about using the UC Libraries, including the electronic resources, see the Library Matters page at the web site. That page also includes information about doing searches of the scientific literature, to find articles on a topic that interests you. Major topic areas there include: UC Berkeley library; electronic journals; journal articles; Medline searches; citation searches.
I no longer list articles that simply announce a new genome sequence. The first organismal (non-viral) sequence was reported in 1995, and was big news. Since then we have has a succession of big announcements (Table 24.1). But now I think that genome sequences per se are no longer news. There are some FR about genome issues, and there are links to genome sites at the web site.
W Martin & M Muller, The hydrogen hypothesis for the first eukaryote. Nature 392:37, 3/5/98. (+ News, Doolittle, p 15.) We broadly understand that prokaryotic and eukaryotic cells are somehow related evolutionarily. In particular, we recognize that the mitochondria and chloroplasts of eukaryotic cells are "prokaryotic-like". Here, based on analysis of biochemical pathways, Martin & Muller propose an alternative view of how the eukaryotic symbiosis arose. They propose that the original bacterial symbiont contributed hydrogen production to the symbiosis -- not respiration as usually assumed. This paper has received much attention, and will guide (provoke?) work to distinguish the old and new models.
A J Gray & A F Raybould, Crop genetics: Reducing transgene escape routes. Nature 392:653, 4/16/98. News. One concern with genetically engineered plants is that the modified gene may be further distributed in nature. Here they discuss work in which the modification is done in the chloroplast genome -- thus preventing spread through pollen.
B Magasanik, A midcentury watershed: the transition from microbial biochemistry to molecular biology. J Bact 181:357, 1/99. A Commentary, part of a series in J Bact to celebrate the centennial anniversary of the American Society for Microbiology.
D C Wallace, Mitochondrial diseases in man and mouse. Science 283:1482, 3/5/99. Review. A variety of degenerative diseases now appear to be due to mitochondrial mutations. Mouse models are beginning to play a role in understanding these diseases.
H N Schulz et al, Dense populations of a giant sulfur bacterium in Namibian shelf deposits. Science 284:493, 4/16/99. (+ News, Wuethrich, p 415.) Thiomargarita namibiensis is now the largest known prokaryote. One Fig in the paper shows one of these bacteria -- along with a fly for comparison. However, these bacteria are mostly vacuole (where they store nutrients for future use). Epulopiscium bacteria are "all meat", with the largest ones almost visible to the naked eye.
K Hopkin, Are we there yet? The Scientist 7/19/99, p 12. What is a "complete" genome sequence? The C elegans sequence was reported as "complete", even though about 1% of the genome remained unsequenced. Because the unsequenced part was deemed hard to sequence, and less interesting, it was common to regard the sequence as "complete". This News article discusses both the serious and amusing aspects of this issue. It is part of a feature issue on Genes and Genomes. For more about The Scientist, including online access, see Sect L.
T J Beveridge, Structures of gram-negative cell walls and their derived membrane vesicles. J Bact 181:4725, 8/99. Minireview. The bacterial cell wall is responsible for the shape, strength, and rigidity of the cell -- yet grows and allows selective transport. Membrane vesicles are an unexpected and poorly understood complexity in bacterial secretion.
Y Michikawa et al, Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 286:774, 10/22/99. They present evidence consistent with the proposal that mutations in mitochondrial DNA are relevant to the aging process.
B Hayes, Computing Science: Experimental Lamarckism. American Scientist 87:494, 11/99. http://bit-player.org/wp-content/extras/bph-publications/AmSci-1999-11-Hayes-Lamarckism.pdf. The now-discredited Lamarckian view of inheritance of acquired characteristics proposed that phenotypic change causes genotypic change. Hayes, a computer scientist, uses computer models to explore when Lamarckian inheritance might be beneficial. Perhaps surprisingly, he shows little benefit of this mode of inheritance.
R Plomin, Genetics and general cognitive ability. Nature 402 Suppl:C25, 12/2/99. An example of genetic studies of complex traits in humans.
C A Hutchison et al, Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286:2165, 12/10/99. (+ related article on ethics, p 2087.) What is the minimum number of genes required for life -- for an independently replicating organism? Well, the smallest gene set identified in nature is the 517 genes of Mycoplasma genitalium. Here, they show that nearly half of these genes appear to be non-essential. That leaves them with about 300 genes that seem to be essential -- including about 100 of unknown function. What makes this article particularly newsworthy is the suggestion that they might try to make an artificial genome containing this minimal set of essential genes. Also see Szostak et al (2001).
P Awadalla et al, Linkage disequilibrium and recombination in hominid mitochondrial DNA. Science 286:2524, 12/24/99. (+ News, Strauss, p 2436.) They suggest that there is some inheritance of mitochondria via the father in humans. Although this has been shown for other organisms, it is thought not to happen in humans. The analysis is indirect, based on observing patterns of mutant sequences, and the conclusion is not completely accepted at this point.
E S Lander & R A Weinberg, Genomics: Journey to the center of biology. Science 287:1777, 3/10/00. Essay; part of the Pathways of Discovery series. A historical view, from Mendel, through the current age of genomics.
M S Fliss et al, Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science 287:2017, 3/17/00. They suggest that detection of mtDNA mutations may be a useful screening method for cancer.
R D Vale and R A Milligan, The way things move: Looking under the hood of molecular motor proteins. Science 288:88-95, 4/7/00. Review. Motor proteins move things, using the energy of ATP hydrolysis to drive the motion. Examples include kinesins, which move chromosomes along microtubules, and the muscle protein myosin, which moves things along actin. Here, they review the emerging details of how the motor molecules function, and discuss the similarities and differences among diverse motor proteins. Among the tools... optical trapping experiments, using laser tweezers, to study the action of single molecules.
D E Gustafson et al, Cryptophyte algae are robbed of their organelles by the marine ciliate Mesodinium rubrum. Nature 405:1049, 6/29/00. Mitochondria and chloroplasts are thought to have arisen by acquisition of prokaryotic cells. Here they show that a photosynthetic protozoan steals chloroplasts from the algae it eats.
P Nurse, The incredible life and times of biological cells. Science 289:1711, 9/8/00. An essay on two centuries of cell biology, by a leading cell biologist, in the Pathways of Discovery series.
H F Willard, Genomics and gene therapy: Artificial chromosomes coming to life. Science 290:1308, 11/17/00. News. Do we understand chromosomes well enough to make artificial chromosomes, by putting together all the pieces? In yeast, yes. Yeast artificial chromosomes (YAC, p 787) are rather well understood and common. However, mammalian artificial chromosomes (MAC) still remain elusive, mainly because of uncertainties about the true nature of the centromere.
A A Diamandopoulos & P C Goudas, Cloning's not a new idea: the Greeks had a word for it centuries ago. Nature 408:905, 12/21/00. Letter. They trace not only the word but the idea of cloning, including cloning humans, back to the time of Aristotle.
J W Szostak et al, Synthesizing life. Nature 409:387, 1/18/01. Part of a set of articles on "unforeseeable science and technology".
1) Q Schiermeier, Testing times for BSE. 2) C Thompson, In search of a cure for CJD. Nature 409:658 & 660, 2/8/01. News features. Useful updates, on practical matters -- testing for and treating prion diseases.
M Fändrich et al, Protein structure: Amyloid fibrils from myoglobin. Nature 410:165, 3/8/01. Neurodegenerative diseases such as Alzheimer's and the prion diseases are characterized, in part, by protein aggregates known as amyloid fibrils. Here they show that myoglobin, an extremely soluble protein with no known propensity to aggregate, will do so under "extreme conditions". Interesting point, in the broad picture of understanding amyloid proteins.
P Chien & J S Weissman, Conformational diversity in a yeast prion dictates its seeding specificity. Nature 410:223, 3/8/01. (+ News, Liebman, p 161.) Prions are infectious agents that lack nucleic acids; they include the agents of the classical scrapie disease in sheep, and BSE. It is now reasonably well accepted that the infectious agent is a protein from a host gene, but with an unusual conformation. Some unusual agents in yeast also seem to behave like prions. The ease of work with microbial systems has allowed rapid progress with "yeast prions" as a model system. One of the confusing issues about prions is the meaning of "strains" -- variants of apparently the same protein with different pathologies. The common view is that strains represent alternative conformations of the prion protein, but some feel that this explanation is unreasonable when there are many strains. Here they explore this issue with yeast prions, and generally support it. A caution... the yeast work can provide clues about what might be, but the relevance to mammalian prions always must remain open.
S Douglas et al, The highly reduced genome of an enslaved algal nucleus. Nature 410:1091, 4/26/01. (+ News, Gilson & McFadden, p 1040.) An oddity, for the curious. Although modern plant chloroplasts are apparently based on a prokaryotic (cyanobacterial) cell, there are some unusual secondary symbioses in which a non-photosynthetic cell has engulfed and retained a eukaryotic algal cell. Here they deal with the highly degenerated nuclear genome of an endosymbiotic red alga. It has 3 chromosomes, 551 kbp, and 531 genes.
L D Hurst, Evolutionary genomics: Sex and the X. Nature 411:149, 5/10/01. News. Discusses recent work on determining the distribution and arrangement of genes. They find that genes for spermatogenesis are over-represented on the Y chromosome, which may seem reasonable, but also on the X.
P V Jallepalli et al, Securin is required for chromosomal stability in human cells. Cell, 105(4):445-457, 5/18/01. Example of work on the fidelity of the mitotic process, with possible relevance to chromosome instability in cancer.
E Szathmary et al, Molecular biology and evolution: Can genes explain biological complexity? Science 292:1315, 5/18/01. A "perspective" article, in the light of the human genome sequencing. They argue that "complexity" may be due more to networking of gene product interactions, than to number of genes.
G P Saborio et al, Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411:810, 6/14/01. A major limitation of the prion theory so far has been the inability to demonstrate that a particular form of the protein is infectious. A key reason for this has been the inability to carry out significant "replication" of the prion (conversion to the disease form) in vitro. Here Saborio et al report what would appear to be a useful replication system. The key advance is actually rather simple: they sonicate the conversion mixture from time to time, thus apparently creating new seeding nuclei. But the critical test remains... is this protein which they converted in vitro infectious? They say that testing is underway. If it is infectious, this will essentially eliminate the remaining objections to the prion theory, and will also open up a variety of new studies, possibly including diagnostics.
A R Weeks et al, A mite species that consists entirely of haploid females. Science 292:2479, 6/29/01. (+ News, Otto & Jarne, p 2441.) The first example of an animal with only haploid females raises questions about the role of diploidy.
Y Gachet et al, A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature 412:352, 7/19/01. (+ News, Nakaseko & Yanagida, p 291.) An interesting recent article on the mechanism of mitosis, more specifically on the control. What prevents sister chromosome separation (anaphase) before the spindle is properly aligned? Here they explore a "checkpoint" -- a step that cannot be passed until certain key things are ready. They show that a regulatory protein senses the proper alignment of the actin cytoskeleton, which itself relates to the spindle.
See the page of Molecular Biology Internet resources. For this chapter, choose Genetics, Cells (Ch 1).
In addition to the links that are listed by chapter, the bottom of the page contains links to sites that focus on a specific organism or specific topic, plus several "miscellaneous" sites.
Also see #7 in the homework set, below.
None reported so far.
Please be alert for possible errors, both in the book and in the handouts. Errors can range from "minor" typos to much bigger problems; all are worth noting. Many errors in the first edition of Weaver were caught by X107 students, and fixed. In fact, students are generally better at catching errors than I am. In general, if you find something that "doesn't seem right", ask about it. You may have found an error, or you may not have understood it right. In the latter case, I can help you with it. Further, I may add a note in the handouts to help clarify the issue.
See Homework section of Syllabus. We will discuss some of the homework in class. The class discussion is best if everyone has worked on the problems, and done as much as you can. Those who want written feedback are welcome to turn in any homework; this may be especially helpful when you miss a homework discussion.
Weaver provides no homework for Ch 1. We need some practice with basic genetics.
1. You have two strains of an ordinary diploid plant. One has red flowers, the other has white flowers. The two strains differ at a single gene.
a. What will be the genotype of the hybrid plants made by crossing these two parents? (Although you may well be able to answer this particular question by inspection, it would be a good exercise to make a gamete matrix.)
b. What color flowers would you expect for these hybrid plants? Explain.
2. Consider a trait determined by a gene with two alleles, B and b. You find a child whose phenotype indicates that s/he is heterozygous for this trait.
a. List the possible sets of genotypes of the parents for this trait?
b. Which of the sets of parental genotypes from part a would produce the highest frequency of heterozygous children?
3. Consider a cross between two bacterial viruses of the genotypes E1S1Y1 and E2S2 Y2.
a. How many gene orders are possible for these three genes? How many if you ignore the direction? Show them. (Remember, at this point you have no information except that there are three genes.)
b. For this part (only), assume that the order is E-S-Y.
i. Sketch the two chromosomes, showing the genes and alleles.
ii. What chromosomes would result if a single crossover event occurred, between the E and S genes?
iii. What chromosomes would result if two crossover events occurred, one between E and S and one between S and Y?
iv. How would the frequencies of the two types of events (last two sub-parts) compare? Why?
Continuing... Now some data:
1000 progeny are examined. 20 are found to be E2S2 Y1, 24 are E2S1Y2, but none of the type E1S2Y2 are found. [The virus is a simple haploid organism, so you need not worry about dominance here. The genes are listed in arbitrary (alphabetical) order; no implication about gene order.]
c. How many types of virus should have resulted from this cross?
d. What is the most likely order of the three genes on the virus chromosome? Explain.
e. Approximately how many of the type E1S1Y2 would you expect were in the progeny? Explain.
f. In the statement of the question, the "genotypes" of the parents are given. Would it have been ok to replace the word "genotypes" with "phenotypes"?
4. Consider two genes which are on the same chromosome but "very far apart" (in genetic terms). How many crossovers are likely to occur between them (e.g., few, many)? What frequency of recombinants will you observe? Why?
5. Even though Weaver does not present much about prokaryotic vs eukaryotic cells, I suspect that many of you are familiar with them. This question should provoke some discussion, both from those who are "expert" in the area and from those who are just learning about the cell types... What are the fundamental differences in functional capabilities between prokaryotic and eukaryotic cells?
6. Improve the chapter summary. If you were going to add one more thought, what would you add? Would you change or delete anything? (This question, which can be considered for any chapter, is a chance to review our perspective on each chapter.)
7. Internet questions.
I will provide occasional questions that ask you to use a resource on the Internet. These problems, of course, are optional, since we are unable to provide you with Internet access. Some of you may have in-house access to some of the tools at work, separate from the Internet. The questions will be simple. The idea is to introduce you to some of the resources. I do not plan to discuss these in class. Feel free to ask me about them privately.
The sites I suggest for you will usually contain many resources; I encourage you to browse around.
The Biology Project at the University of Arizona maintains a web site that covers a broad range of biological topics. For each topic, practice questions, with good feedback, and tutorials are available. As a start, go to
http://www.biology.arizona.edu/
and choose Mendelian Genetics, for some practice. But also look through the list of topics, for future reference.
I would appreciate feedback, pro or con, if you try this site.
1. a. One strain is homozygous for 'redness'; let's call its genotype RR. The other strain is homozygous for 'whiteness'; genotype WW. (How do you know the given strains are homozygous? It's implicit. If they weren't, they wouldn't breed true, and we wouldn't refer to them as strains.)
The genotype of the hybrid is clear enough: they will all be RW heterozygotes, according to Mendel's principle of independent segregation.
A gamete matrix to show this would look something like the following. Expected gametes from the two parents are across the top and along the left side. Each internal cell is based on its row and column gametes; each such cell shows the genotype of one portion of the progeny.
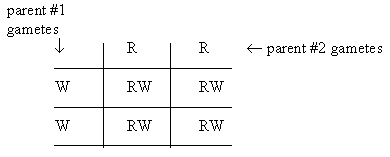
Each of the 4 internal cells represents 25% of the progeny, since each gamete type is equally likely. In this case, each internal cell is RW, the genotype of all hybrids from this cross.
b. As to the phenotype, there is absolutely no way to know, based on the given information. There is no information that allows you to predict which allele -- if either -- is dominant. One finds which is dominant by observing the phenotype of the hybrid (or, sometimes, by knowing the function of the individual alleles).
If you said that the hybrids would be all red, you made an assumption (that red is dominant) not based on any given information.
On a test, it is important to explicitly state any assumptions that you make. Sometimes it is necessary or useful to make assumptions, other times it's preferable that you stay within the given information. But if you state your assumptions I will be better able to follow your line of thinking. This may well lead to more points; it should at least lead to better feedback. To use this case as an example, if you just say that the hybrid flowers would be all red, that would probably earn a zero. If you say they would be all red because you assumed that red is dominant, you have indicated that you noticed the lack of dominance information in the question. I would prefer another assumption (dominance explicitly unknown), but your clearly explained answer would probably earn credit -- plus a note focusing on your assumption.
2. a. BB x Bb, BB x bb, Bb x Bb, Bb x bb. In fact, most possible combinations would produce some heterozygotes, except which ones???
b. One of those combinations will produce 100% heterozygotes. Which one?
3. a. 6; 3. If you ignore direction, the only distinction would be which gene is in the middle. There are three genes, so there are three possibilities for whom is in the middle.
b. i.
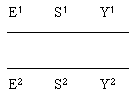
ii. E1S2Y2 and E2 S1Y1
iii. E1S2Y1 and E2 S1Y2
iv. The event is part ii is more frequent because it only requires one crossover.
c. There are three genes, each with two alleles. Thus there are 23 (=8) possible types (though at least one type had 0 number in this case).
d. The order is YES (or SEY; there is no information on direction). The low frequency (absence) of one class indicates that it requires a double crossover; therefore E must be in the middle. [Expected number for the double crossover class is (2 x 2%) x (2 x 2.4%) / 2 = 0.1% = 1 out of 1000. Observed 0.]
e. 20 (out of 1000), same as the reciprocal recombinant, E2S2 Y1.
4. Many crossovers will occur. That's what we mean by far apart. What is the consequence for the observed result of percent recombinants? Think about... what if there is an odd number of crossovers? An even number?
Molecular biology chapter handouts information Molecular biology home page
Contact information Site home page
Last update: June 3, 2019